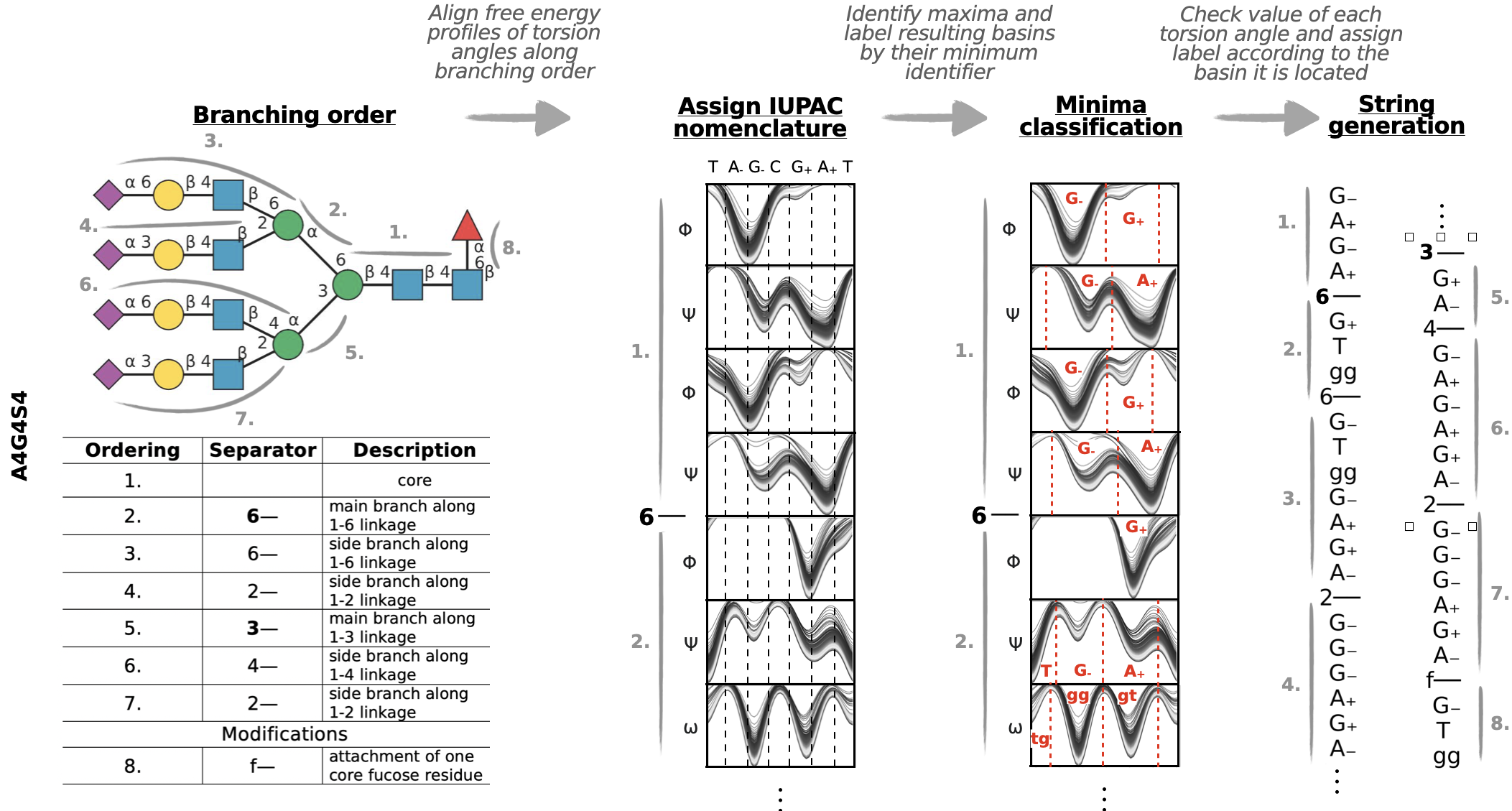
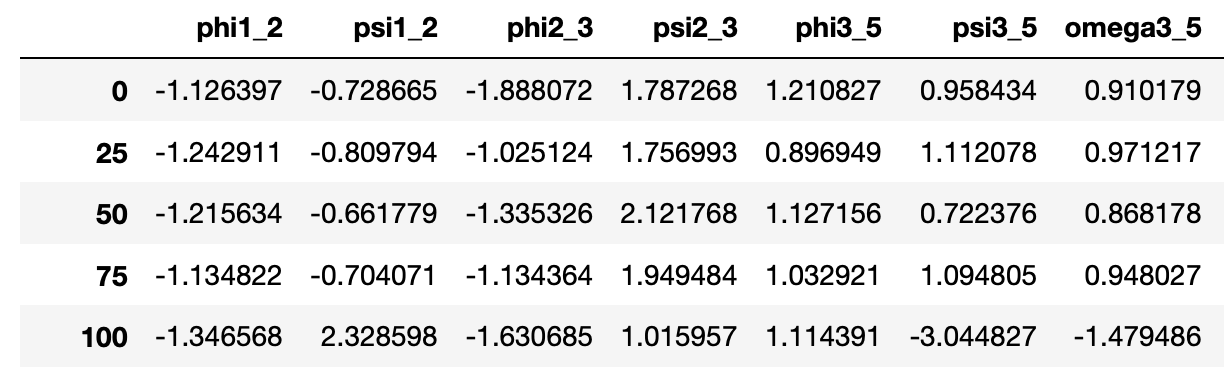
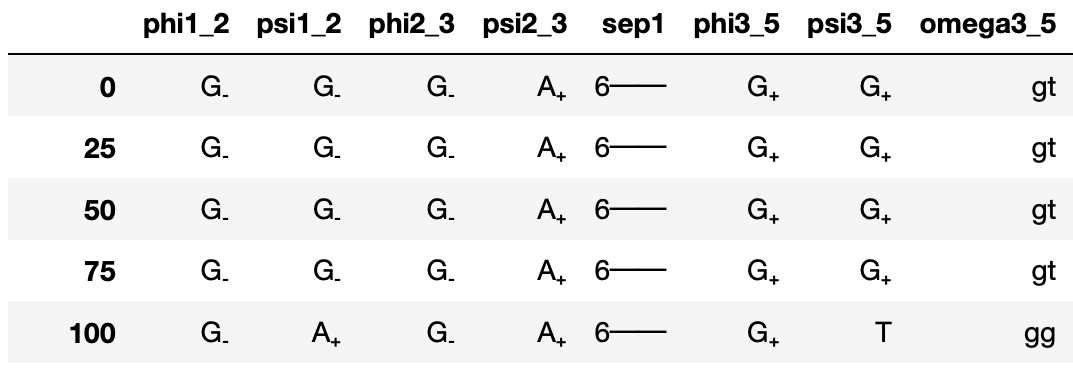
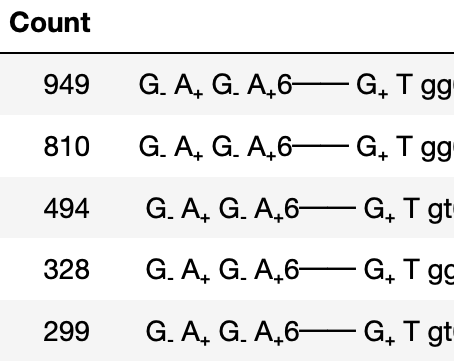
GlyCONFORMERS is a Python package that assigns conformer strings to N-glycan conformers, based on their torsion angle values.
Project description
GlyCONFORMER
GlyCONFORMERS is a Python package that assigns labels to glycan conformers, based on their torsion angle values, in order to differentiate various 3D-structures of one single glycan. It enables the automated assignment of the GlyCONFORMER string that was introduced in:
Grothaus et al. 2022, Exploration, Representation, and Rationalization of the Conformational Phase Space of N-Glycans, J. Chem. Inf. Model. 2022, 62, 20, 4992–5008 https://pubs.acs.org/doi/full/10.1021/acs.jcim.2c01049 for N-glycans.
Check the paper or tutorial for a detailed explanation of the GlyCONFORMER string generation. The workflow is not exclusivly designed for N-glycans, but can also be applyied to any other glycan type there is.
Installation
GlyCONFORMER is available via pip and docker:
pip
pip install GlyCONFORMER
Stable performance was only tested and verified up to python version 3.10. Pandas<=1.3.5 is required!
Recommendation - build via conda environment with following commands:
conda create -n name python=3.10 --no-default-packages
conda activate name
pip install GlyCONFORMER
Docker Image
This image includes:
- GlyCONFORMER pre-installed via
pip install . - Jupyter Notebook launched on container start
- Required dependencies like
plumed,numpy,scikit-learn, etc.
docker pull grothaus/glyconformer:latest
docker run -p 8888:8888 grothaus/glyconformer:latest
Tutorial
The tutorial includes different N-glycan types and different complexity levels of how to obtain a GlyCONFORMER label string for custom glycan types and their recorded torsion angle values. The minimum example is given by the high-mannose type N-glycan M5, where all necessary information are read from the LIBRARY_GLYCANS folder by specifying the glycantype = "Man5":
M5 = Glyconformer(glycantype = "Man5")
Raw torsion angle values get converted to letters corresponding to their values and associate them to a certain minima of the free energy profile along that torsion angle.
The raw input data can be accessed via:
M5.colvar
and its translated variant via:
M5.binary
Additionally, the occurance of each conformer string is counted and outputted via:
M5.count
All this is performed in the background and final results can be outputted using various plotting functions, e.g.:
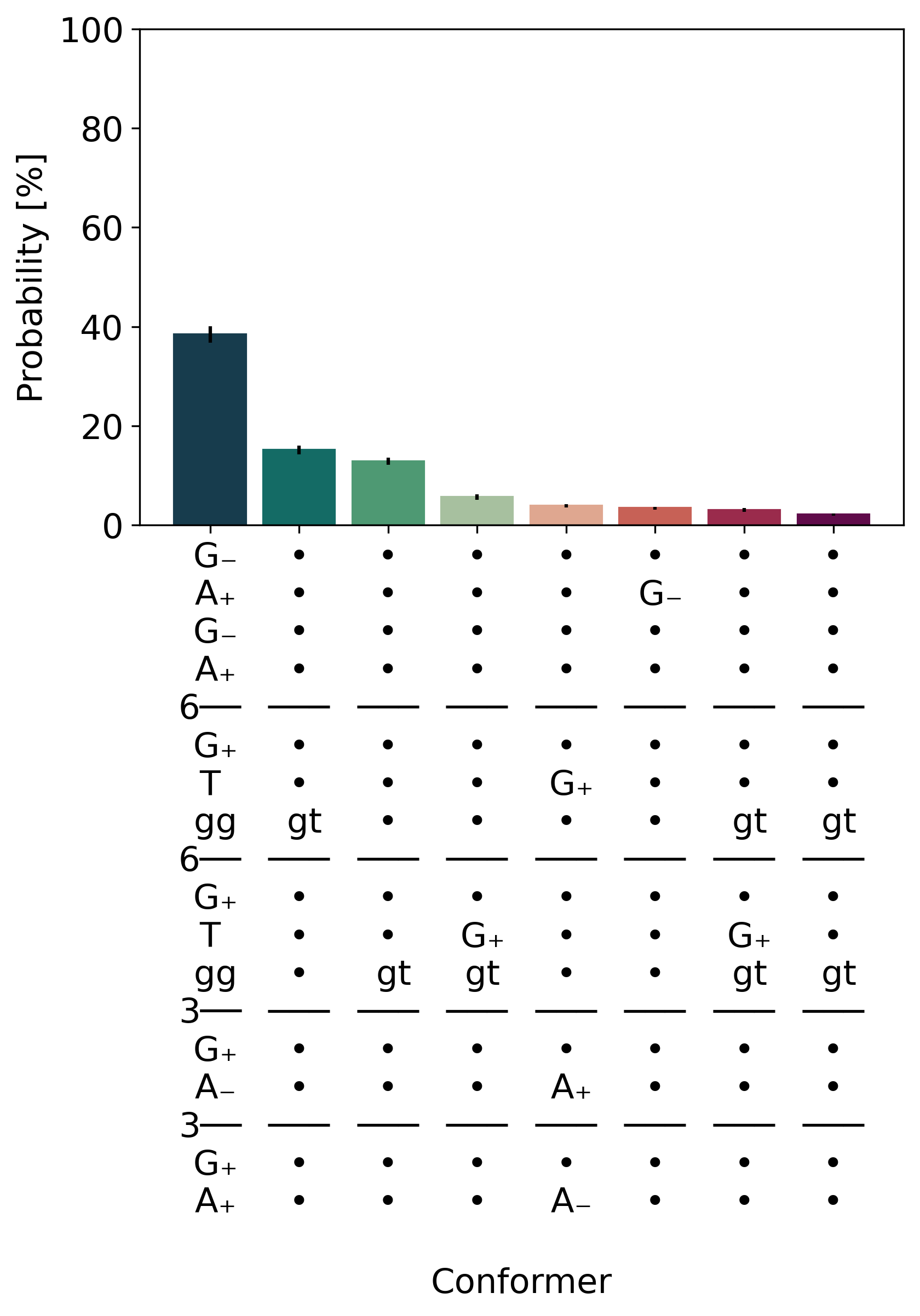
M5.distribution
Conformer labels are given on the x-axis and deviations from the most populated conformer indicated by explicit letters, where dots are used when no change in that torsion angle could be detected.
For more elaborate examples and a detailed explanation of how free energy profiles of each torsion angle are classified, check out the Tutorial_GlyCONFORMER.ipynb notebook.
Documentation
See documentation https://glyconformer.readthedocs.io/en/latest/index.html
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file GlyCONFORMER-1.1.3.tar.gz.
File metadata
- Download URL: GlyCONFORMER-1.1.3.tar.gz
- Upload date:
- Size: 30.6 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.7.12
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
c191e71b004d0e897c697d95bfdd2ee797fa4b6b6588aa1e11d43c4e7b697a47
|
|
| MD5 |
22b98b875766b85837db54955f8414e3
|
|
| BLAKE2b-256 |
bfa612855ef3f3bac252fb08b1e9b361c35bee7dafef01d3574f7d65c8efbd9b
|
File details
Details for the file GlyCONFORMER-1.1.3-py3-none-any.whl.
File metadata
- Download URL: GlyCONFORMER-1.1.3-py3-none-any.whl
- Upload date:
- Size: 29.5 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.7.12
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
16b9dafe4683b138d31e21f8a4c4b6169e37c75543da4ac8743101cf17804756
|
|
| MD5 |
407a92a94476a2f013b4a4239af74ea8
|
|
| BLAKE2b-256 |
5e97a1022d1253205a6bc9f16afcdf50a8e96dfe29fc09aa9d8a009f36fde3f3
|