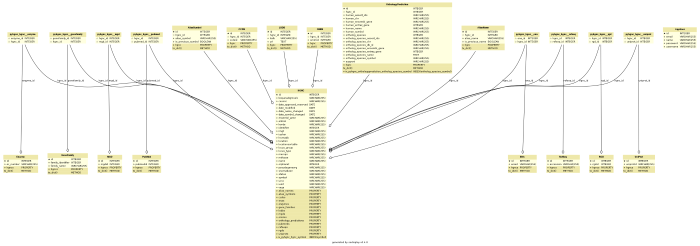
Importing and querying HGNC data
Project description
PyHGNC is a Python package to access and query data provided by HGNC-approved gene nomenclature, gene families and associated resources including links to genomic, proteomic and phenotypic information.
Data are installed in a (local or remote) RDBMS enabling bioinformatic algorithms very fast response times to sophisticated queries and high flexibility by using SOLAlchemy database layer.
PyHGNC is developed by the Department of Bioinformatics at the Fraunhofer Institute for Algorithms and Scientific Computing SCAI For more in for information about PyHGNC go to the documentation.
This development is supported by following IMI projects:
Supported databases
PyHGNC uses SQLAlchemy to cover a wide spectrum of RDMSs (Relational database management system). For best performance MySQL or MariaDB is recommended. But if you have no possibility to install software on your system SQLite - which needs no further installation - also works. Following RDMSs are supported (by SQLAlchemy):
Getting Started
This is a quick start tutorial for impatient.
Installation

PyHGNC can be installed with pip.
pip install pyhgncIf you fail because you have no rights to install use superuser (sudo on Linux before the commend) or …
pip install --user pyhgncIf you want to make sure you are installing this under python3 use …
python3 -m pip install pyhgncSQLite
If you don’t know what all that means skip the section MySQL/MariaDB setup.
Don’t worry! You can always later change the configuration. For more information about changing database system later go to the subtitle Changing database configuration Changing database configuration in the documentation on readthedocs.
MySQL/MariaDB setup
Log in MySQL as root user and create a new database, create a user, assign the rights and flush privileges.
CREATE DATABASE pyhgnc CHARACTER SET utf8 COLLATE utf8_general_ci;
GRANT ALL PRIVILEGES ON pyhgnc.* TO 'pyhgnc_user'@'%' IDENTIFIED BY 'pyhgnc_passwd';
FLUSH PRIVILEGES;There are two options to set the MySQL/MariaDB.
The simplest is to start the command line tool
pyhgnc mysqlYou will be guided with input prompts. Accept the default value in squared brackets with RETURN. You will see something like this
server name/ IP address database is hosted [localhost]:
MySQL/MariaDB user [pyhgnc_user]:
MySQL/MariaDB password [pyhgnc_passwd]:
database name [pyhgnc]:
character set [utf8]:Connection will be tested and in case of success return Connection was successful. Otherwise you will see following hint
Test was NOT successful
Please use one of the following connection schemas
MySQL/MariaDB (strongly recommended):
mysql+pymysql://user:passwd@localhost/database?charset=utf8
PostgreSQL:
postgresql://user:passwd@localhost/database
MsSQL (pyodbc needed):
mssql+pyodbc://user:passwd@database
SQLite (always works):
- Linux:
sqlite:////absolute/path/to/database.db
- Windows:
sqlite:///C:\absolute\path\to\database.db
Oracle:
oracle://user:passwd@localhost:1521/database2. The second option is to start a python shell and set the MySQL configuration. If you have not changed anything in the SQL statements above …
import pyhgnc
pyhgnc.set_mysql_connection()If you have used you own settings, please adapt the following command to you requirements.
import pyhgnc
pyhgnc.set_mysql_connection(host='localhost', user='pyhgnc_user', passwd='pyhgnc_passwd', db='pyhgnc')Updating
The updating process will download the complete HGNC json file and the HCOP file.
import pyhgnc
pyhgnc.manager.database.update()This will use either the default connection settings of PyHGNC or the settings defined by the user. It is also possible to run the update process from shell.
pyhgnc updateQuick start with query functions
Initialize the query object
query = pyhgnc.query()Get all HGNC entries:
all_entries = query.hgnc()More information
See the installation documentation for more advanced
instructions. Also, check the change log at CHANGELOG.rst.
HGNC tools
HGNC provides also online tools .
Links
HUGO Gene Nomenclature Committee (HGNC)
PyHGNC
Documented on Read the Docs
Versioned on GitHub
Tested on Travis CI
Distributed by PyPI
Chat on Gitter
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distributions
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file PyHGNC-0.2.3.tar.gz.
File metadata
- Download URL: PyHGNC-0.2.3.tar.gz
- Upload date:
- Size: 494.3 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
910f9a55c7b378a9a782fbfcbfe585fb7ad6ba5a50b9e7086184c2a45722915f
|
|
| MD5 |
c263cb1b25ecb5a77a567b30e46476f9
|
|
| BLAKE2b-256 |
7e647d34b4decb7414a5fca7742cd81ba8e4eed0c4d0d9f9035ae5b2f1fb817f
|
File details
Details for the file PyHGNC-0.2.3-py3-none-any.whl.
File metadata
- Download URL: PyHGNC-0.2.3-py3-none-any.whl
- Upload date:
- Size: 32.5 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
0b196a53119f6193a94d9a815dc62ac7d7ac8e96f5633528e4ec88a2394a4a97
|
|
| MD5 |
31f678651851d855c54cb3c2afbef886
|
|
| BLAKE2b-256 |
b922cd606c448d09b712cb1a7061994e15563a149406393747a08a418088c433
|
File details
Details for the file PyHGNC-0.2.3-py2-none-any.whl.
File metadata
- Download URL: PyHGNC-0.2.3-py2-none-any.whl
- Upload date:
- Size: 32.5 kB
- Tags: Python 2
- Uploaded using Trusted Publishing? No
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
88b3910f6300ce869a15483b69fb5db4ffd4ad014a0753c067e3eb65ee96dd1f
|
|
| MD5 |
03177a929b96bb4db9d9d74117198ec0
|
|
| BLAKE2b-256 |
d2240e5588f0a12d5129554334eb5def720a19f632116055170db6c0bb8a8b14
|