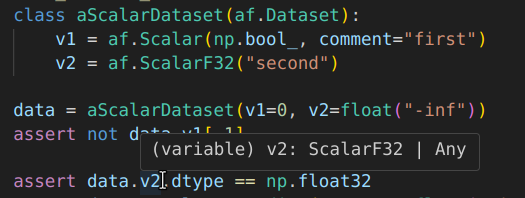
Module for creating well-documented datasets, with types and annotations.
Project description
Affinity
Affinity makes it easy to create well-annotated datasets from vector data. What your data means should always travel together with the data.
Affinity is a pythonic dialect of Data Definition Language (DDL). Affinity does not replace any dataframe library, but can be used with any package you like.
If you're unsatisfied that documenting your data models has remained an afterthought, check out the ideas here.
Installation
Install with any flavor of pip install affinity, or copy affinity.py into your project. It's only one file.
🐼 🦆 Affinity requires Pandas (works with v2 and v3) and DuckDB (1.3 and up). Polars and pyarrow are optional.
Usage
Now all your data models can be concisely declared as python classes.
import affinity as af
class SensorData(af.Dataset):
"""Experimental data from Top Secret Sensor Tech."""
t = af.VectorF32("elapsed time (sec)")
channel = af.VectorI8("channel number (left to right)")
voltage = af.VectorF64("something we measured (mV)")
is_laser_on = af.VectorBool("are the lights on?")
exp_id = af.ScalarI32("FK to `experiment`")
LOCATION = af.Location(folder="s3://mybucket/affinity", file="raw.parquet", partition_by=["channel"])
# how to use affinity Datasets:
data = SensorData() # ✅ empty dataset
data = SensorData(**fields) # ✅ build manually
data = SensorData.build(...) # ✅ build from a source (dataframes, DuckDB)
data.df # .pl / .arrow # ✅ view as dataframe (Pandas/Polars/Arrow)
data.metadata # ✅ annotations (data dict with column and dataset comments), origin
data.origin # ✅ creation metadata, some data provenance
data.sql(...) # ✅ run DuckDB SQL query on the dataset
data.to_parquet(...) # ✅ data.metadata -> Parquet metadata
data.partition() # ✅ get formatted paths and partitioned datasets
How it works
The af.Dataset is Affinity's BaseModel, the base class that defines the behavior of children data classes:
- concise class declaration sets the expected dtypes and descriptions for each attribute (column)
- class attributes can be represented by any array (defaults to
pd.Seriesbecause it handles nullable integers well) - class instances can be constructed from scalars, vectors/iterables, or other datasets
- type hints for scalar and vector data
Detailed example: Parquet Round-Trip
All you need to create a data class are typed classes and comments explaining what the fields mean.
1. Declare class
import affinity as af
class IsotopeData(af.Dataset):
"""NIST Atomic Weights & Isotopic Compositions.[^1]
[^1] https://www.nist.gov/pml/atomic-weights-and-isotopic-compositions-relative-atomic-masses
"""
symbol = af.VectorObject("Element")
z = af.VectorI8("Atomic Number (Z)")
mass = af.VectorF64("Isotope Mass (Da)")
abundance = af.VectorF64("Relative natural abundance")
IsotopeData.z
# DescriptorType Int8 of len 0 # Atomic Number (Z)
# Series([], dtype: Int8)
IsotopeData().pl # show fields and types
# shape: (0, 4)
# symbol z mass abundance
# str i8 f64 f64
IsotopeData.LOCATION # new in v0.4
# Location(folder=PosixPath('.'), file='IsotopeData_export.csv', partition_by=[])
The class attributes are instantiated Vector objects of zero length. Using the descriptor pattern, they are replaced with actual data arrays on building the instance.
2. Build class instance from querying a CSV
To build the dataset, we use IsotopeData.build() method with query argument. We use DuckDB FROM-first syntax, with rename=True keyword argument. The fields in the query result will be assigned names and types provided in the class definition. With rename=False (default), the source columns must be named exactly as class attributes. When safe type casting is not possible, an error will be raised; element with z=128 would not fit this dataset. Good thing there isn't one (not even as a Wikipedia article)!
url = "https://raw.githubusercontent.com/liquidcarbon/chembiodata/main/isotopes.csv"
data_from_sql = IsotopeData.build(query=f"FROM '{url}'", rename=True)
# data_from_sql = IsotopeData.build(query=f"FROM '{url}'") # will fail
query_without_rename = f"""
SELECT
Symbol as symbol,
Number as z,
Mass as mass,
Abundance as abundance,
FROM '{url}'
"""
data_from_sql2 = IsotopeData.build(query=query_without_rename)
assert data_from_sql == data_from_sql2
print(data_from_sql)
# Dataset IsotopeData of shape (354, 4)
# symbol = ['H', 'H' ... 'Ts', 'Og']
# z = [1, 1 ... 117, 118]
# mass = [1.007825, 2.014102 ... 292.20746, 294.21392]
# abundance = [0.999885, 0.000115 ... 0.0, 0.0]
3. Write to Parquet, with metadata.
data_from_sql.to_parquet("test.parquet") # requires PyArrow
4. Inspect metadata using PyArrow:
The schema metadata as shown here is truncated; full-length keys and values are in pf.schema_arrow.metadata.
import pyarrow.parquet as pq
pf = pq.ParquetFile("isotopes.parquet")
pf.schema_arrow
# symbol: string
# z: int8
# mass: double
# abundance: double
# -- schema metadata --
# table_comment: 'NIST Atomic Weights & Isotopic Compositions.[^1]
# [' + 97
# symbol: 'Element'
# z: 'Atomic Number (Z)'
# mass: 'Isotope Mass (Da)'
# abundance: 'Relative natural abundance'
# created_ts: '1724787055721'
# source: 'dataframe, shape (354, 4)
# query:
# SELECT
# Symbol as symbol,
# ' + 146
[!TIP] Though in all examples here the comment field is a string, Arrow allows non-string data in Parquet metadata (some caveats apply). If you're packaging multidimensional vectors, check out "test_objects_as_metadata" in the test file.
5. Inspect metadata using DuckDB
DuckDB provides several functions for querying Parquet metadata. We're specifically interested in key-value metadata, where both keys and values are of type BLOB. It can be decoded on the fly using SELECT DECODE(key), DECODE(value) FROM parquet_kv_metadata(...), or like so:
import duckdb
source = duckdb.sql("FROM parquet_kv_metadata('isotopes.parquet') WHERE key='source'")
print(source.fetchall()[-1][-1].decode())
# dataframe, shape (354, 4)
# query:
# SELECT
# Symbol as symbol,
# Number as z,
# Mass as mass,
# Abundance as abundance,
# FROM 'https://raw.githubusercontent.com/liquidcarbon/chembiodata/main/isotopes.csv'
6. Read Parquet:
data_from_parquet = IsotopeData.build(query="FROM 'isotopes.parquet'")
assert data_from_sql == data_from_parquet
print(data_from_parquet.pl.dtypes)
# [String, Int8, Float64, Float64]
7. Bonus: Partitions
The special attribute LOCATION helps you write the data where you want, how you want it. LOCATION does not have to be declared, but it is set to sensible (unpartitioned) defaults.
On calling af.Dataset.partition(), you'll get the formatted list of Hive-style partitions and the datasets broken up accordingly.
This is en route to af.Dataset.save(), which in all likelihood won't be done since there's far too many ways to handle this.
class PartitionedIsotopeData(af.Dataset):
symbol = af.VectorObject("Element")
z = af.VectorI8("Atomic Number (Z)")
mass = af.VectorF64("Isotope Mass (Da)")
abundance = af.VectorF64("Relative natural abundance")
LOCATION = af.Location(folder="s3://myisotopes", file="data.csv", partition_by=["z"])
url = "https://raw.githubusercontent.com/liquidcarbon/chembiodata/main/isotopes.csv"
data_from_sql = PartitionedIsotopeData.build(query=f"FROM '{url}'", rename=True)
names, folders, filepaths, datasets = data_from_sql.partition()
# this variety of outputs is helpful when populating cloud warehouses,
# such as Athena/Glue via awswrangler.
names[:3], folders[:3]
# ([['1'], ['2'], ['3']], ['s3://myisotopes/z=1/', 's3://myisotopes/z=2/', 's3://myisotopes/z=3/'])
#
filepaths[:3], datasets[:3]
# (['s3://myisotopes/z=1/data.csv', 's3://myisotopes/z=2/data.csv', 's3://myisotopes/z=3/data.csv'], [Dataset PartitionedIsotopeData of shape (3, 4)
# symbol = ['H', 'H', 'H']
# z = [1, 1, 1]
# mass = [1.007825, 2.014102, 3.016049]
# abundance = [0.999885, 0.000115, 0.0], Dataset PartitionedIsotopeData of shape (2, 4)
# symbol = ['He', 'He']
# z = [2, 2]
# mass = [3.016029, 4.002603]
# abundance = [1e-06, 0.999999], Dataset PartitionedIsotopeData of shape (2, 4)
# symbol = ['Li', 'Li']
# z = [3, 3]
# mass = [6.015123, 7.016003]
# abundance = [0.0759, 0.9241]])
If you work with AWS Athena, also check out kwargs_for_create_athena_table method available on all Datasets.
Motivation
Once upon a time, relational databases met object-oriented programming, and gave rise to object-relational mapping. Django ORM and SQLAlchemy unlocked the ability to represent database entries as python objects, with attributes for columns and relations, and methods for create-read-update-delete (CRUD) transactions. These scalar data elements (numbers, strings, booleans) carry a lot of meaning: someone's name or email or account balance, amounts of available items, time of important events. They change relatively frequently, one row at a time, and live in active, fast memory (RAM).
future blurb about OLAP and columnar and cloud data storage
We need something new for vector data.
There are many options for working with dataframes composed of vectors - pandas, polars, pyarrow are all excellent - there are a few important pieces missing:
- other than variable and attribute names, there's no good way to explain what the dataset and each field is about; what the data means is separated from the data itself
- dataframe packages are built for maximum flexibility in working with any data types; this leads to data quality surprises and is sub-optimal for storage and compute
Consider the CREATE TABLE statement in AWS Athena, the equivalent of which does not exist in any one python package:
CREATE EXTERNAL TABLE [IF NOT EXISTS]
[db_name.]table_name [(col_name data_type [COMMENT col_comment] [, ...] )]
[COMMENT table_comment]
[PARTITIONED BY (col_name data_type [COMMENT col_comment], ...)]
[CLUSTERED BY (col_name, col_name, ...) INTO num_buckets BUCKETS]
[ROW FORMAT row_format]
[STORED AS file_format]
[WITH SERDEPROPERTIES (...)]
[LOCATION 's3://amzn-s3-demo-bucket/[folder]/']
[TBLPROPERTIES ( ['has_encrypted_data'='true | false',] ['classification'='aws_glue_classification',] property_name=property_value [, ...] ) ]
Affinity exists to fill (some of) these gaps.
Tales of Data Annotation Issues Gone Terribly Wrong
Probably the single greatest source of problems is unspecified units of measure, with numerous fatal and near-fatal engineering and medical disasters.
Have you ever stared at a bunch of numbers and had no clue what they represented? Do you have an anecdote of bad things happening due un/misannotated data? Share in discussions!
Future
- nested data - WIP, but this already works:
# nested datasets serialize as dicts(structs)
import affinity as af
class User(af.Dataset):
name = af.ScalarObject("username")
attrs = af.VectorObject("user attributes")
class Task(af.Dataset):
created_ts = af.ScalarF64("created timestamp")
user = User.as_field("vector")
hours = af.VectorI16("time worked (hours)")
u1 = User(name="Alice", attrs=["adorable", "agreeable"])
u2 = User(name="Brent", attrs=["bland", "broke"])
t1 = Task(created_ts=123.456, user=[u1, u2], hours=[3, 5])
t1.to_parquet("task.parquet")
duckdb.sql("FROM 'task.parquet'")
# ┌────────────┬─────────────────────────────────────────────────┬───────┐
# │ created_ts │ user │ hours │
# │ double │ struct(attrs varchar[], "name" varchar) │ int16 │
# ├────────────┼─────────────────────────────────────────────────┼───────┤
# │ 123.456 │ {'attrs': [adorable, agreeable], 'name': Alice} │ 3 │
# │ 123.456 │ {'attrs': [bland, broke], 'name': Brent} │ 5 │
# └────────────┴─────────────────────────────────────────────────┴───────┘
# return flatted dataframe
t1.flatten(prefix=True) # unnested columns are prefixed (user.name, user.attrs)
t1.flatten(prefix=False) # default: keep original column names (name, attrs)
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file affinity-1.1.0.tar.gz.
File metadata
- Download URL: affinity-1.1.0.tar.gz
- Upload date:
- Size: 11.1 kB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.0.1 CPython/3.12.8
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
1b941d6be5b9d6c9916e88aff0020493e5b80b73f9406b2a8ed399ba9506d1dd
|
|
| MD5 |
4641e6c43182c3b82f00c86ff2d408ff
|
|
| BLAKE2b-256 |
46e1e4bc611b46cea13bca73dbee33e4ea0431b46bcf2e03f1c75c0302cb5e1e
|
Provenance
The following attestation bundles were made for affinity-1.1.0.tar.gz:
Publisher:
publish_pypi.yml on liquidcarbon/affinity
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
affinity-1.1.0.tar.gz -
Subject digest:
1b941d6be5b9d6c9916e88aff0020493e5b80b73f9406b2a8ed399ba9506d1dd - Sigstore transparency entry: 872464100
- Sigstore integration time:
-
Permalink:
liquidcarbon/affinity@682166784523edd3314a48e5528d2299499b1976 -
Branch / Tag:
refs/heads/main - Owner: https://github.com/liquidcarbon
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
publish_pypi.yml@682166784523edd3314a48e5528d2299499b1976 -
Trigger Event:
workflow_run
-
Statement type:
File details
Details for the file affinity-1.1.0-py3-none-any.whl.
File metadata
- Download URL: affinity-1.1.0-py3-none-any.whl
- Upload date:
- Size: 11.5 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.0.1 CPython/3.12.8
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
10436256c954ef717cd2d620c245b2724f31e7b37fa42b1511ca9b3f7d330389
|
|
| MD5 |
9c5add48a38c80075fc03cd5c63792a2
|
|
| BLAKE2b-256 |
1e5f022226e26d7b9f8f0c6f4eb80d74e8de7031236e0dd33a866fb3510292a4
|
Provenance
The following attestation bundles were made for affinity-1.1.0-py3-none-any.whl:
Publisher:
publish_pypi.yml on liquidcarbon/affinity
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
affinity-1.1.0-py3-none-any.whl -
Subject digest:
10436256c954ef717cd2d620c245b2724f31e7b37fa42b1511ca9b3f7d330389 - Sigstore transparency entry: 872464103
- Sigstore integration time:
-
Permalink:
liquidcarbon/affinity@682166784523edd3314a48e5528d2299499b1976 -
Branch / Tag:
refs/heads/main - Owner: https://github.com/liquidcarbon
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
publish_pypi.yml@682166784523edd3314a48e5528d2299499b1976 -
Trigger Event:
workflow_run
-
Statement type: