Jigsaw-like AggMap: A Robust and Explainable Omics Deep Learning Tool
Project description

Jigsaw-like AggMap
A Robust and Explainable Omics Deep Learning Tool
Installation
install aggmap by:
# create an aggmap env
conda create -n aggmap python=3.8
conda activate aggmap
pip install --upgrade pip
pip install aggmap
Usage
import pandas as pd
from sklearn.datasets import load_breast_cancer
from aggmap import AggMap, AggMapNet
# Data loading
data = load_breast_cancer()
dfx = pd.DataFrame(data.data, columns=data.feature_names)
dfy = pd.get_dummies(pd.Series(data.target))
# AggMap object definition, fitting, and saving
mp = AggMap(dfx, metric = 'correlation')
mp.fit(cluster_channels=5, emb_method = 'umap', verbose=0)
mp.save('agg.mp')
# AggMap visulizations: Hierarchical tree, embeddng scatter and grid
mp.plot_tree()
mp.plot_scatter(enabled_data_labels=True, radius=5)
mp.plot_grid(enabled_data_labels=True)
# Transoformation of 1d vectors to 3D Fmaps (-1, w, h, c) by AggMap
X = mp.batch_transform(dfx.values, n_jobs=4, scale_method = 'minmax')
y = dfy.values
# AggMapNet training, validation, early stopping, and saving
clf = AggMapNet.MultiClassEstimator(epochs=50, gpuid=0)
clf.fit(X, y, X_valid=None, y_valid=None)
clf.save_model('agg.model')
# Model explaination by simply-explainer: global, local
simp_explainer = AggMapNet.simply_explainer(clf, mp)
global_simp_importance = simp_explainer.global_explain(clf.X_, clf.y_)
local_simp_importance = simp_explainer.local_explain(clf.X_[[0]], clf.y_[[0]])
# Model explaination by shapley-explainer: global, local
shap_explainer = AggMapNet.shapley_explainer(clf, mp)
global_shap_importance = shap_explainer.global_explain(clf.X_)
local_shap_importance = shap_explainer.local_explain(clf.X_[[0]])
How It Works?
- AggMap flowchart of feature mapping and agglomeration into ordered (spatially correlated) multi-channel feature maps (Fmaps)
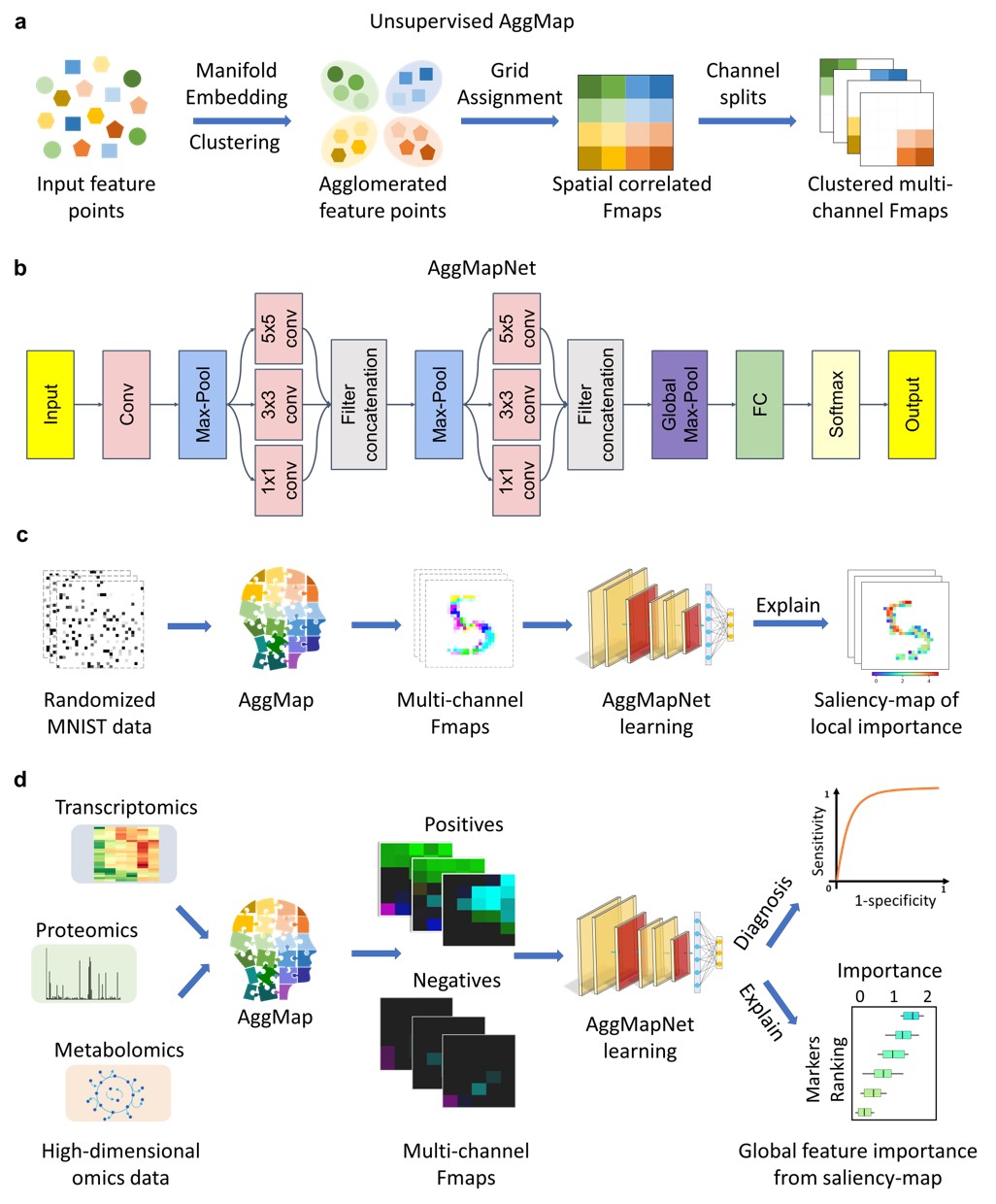
saliency-map of important features).
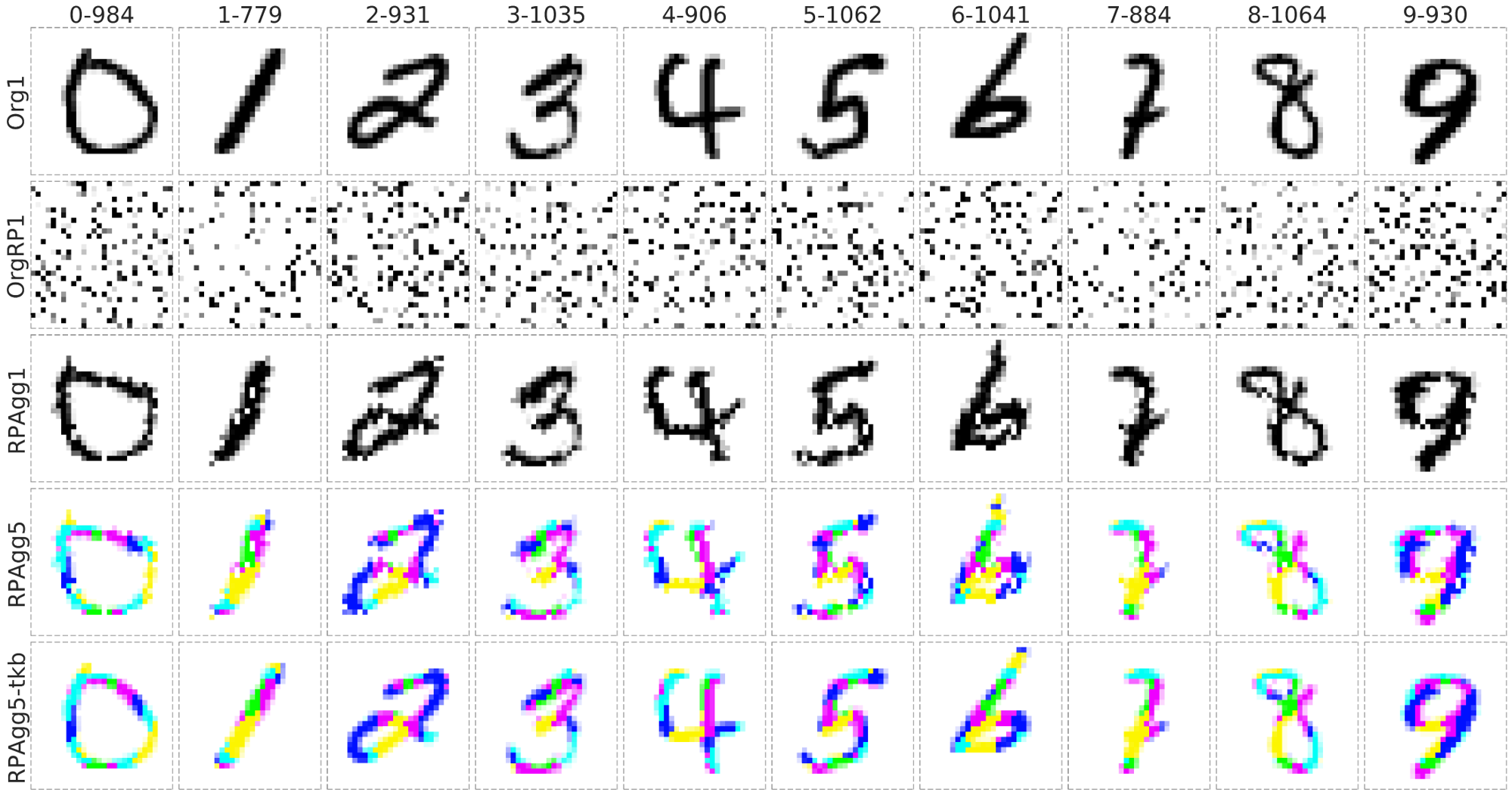
Proof-of-Concepts of reconstruction ability on MNIST Dataset
- It can reconstruct to the original image from completely randomly permuted (disrupted) MNIST data:
Org1: the original grayscale images (channel = 1), OrgRP1: the randomized images of Org1 (channel = 1), RPAgg1, 5: the reconstructed images of OrgPR1 by AggMap feature restructuring (channel = 1, 5 respectively, each color represents features of one channel). RPAgg5-tkb: the original images with the pixels divided into 5 groups according to the 5-channels of RPAgg5 and colored in the same way as RPAgg5.
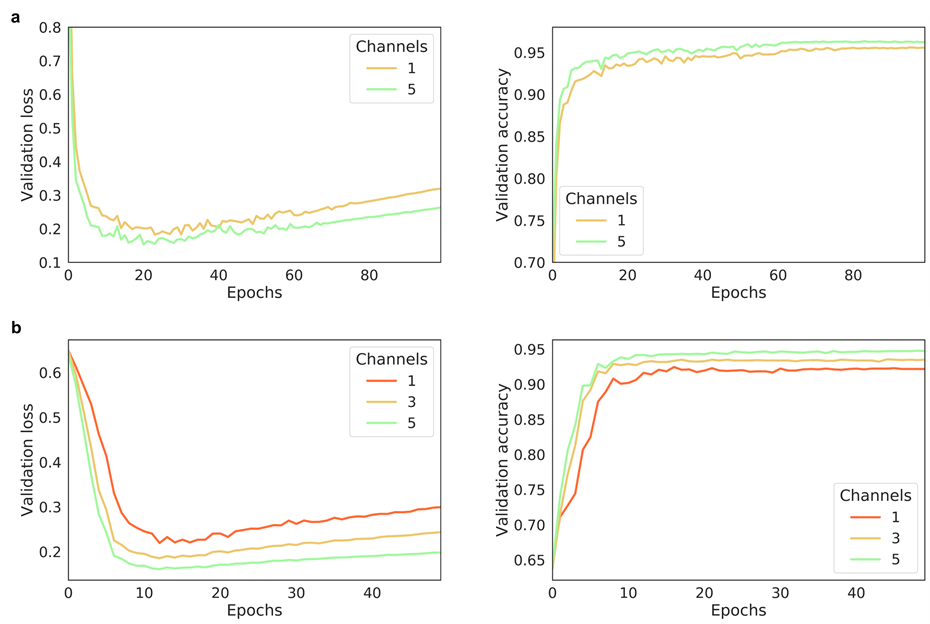
The effect of the number of channels on model performance
- Multi-channel Fmaps can boost the model performance notably:
The performance of AggMapNet using different number of channels on the TCGA-T (a) and COV-D (b). For TCGA-T, ten-fold cross validation average performance, for COV-D, a fivefold cross validation was performed and repeat 5 rounds using different random seeds (total 25 training times), their average performances of the validation set were reported.
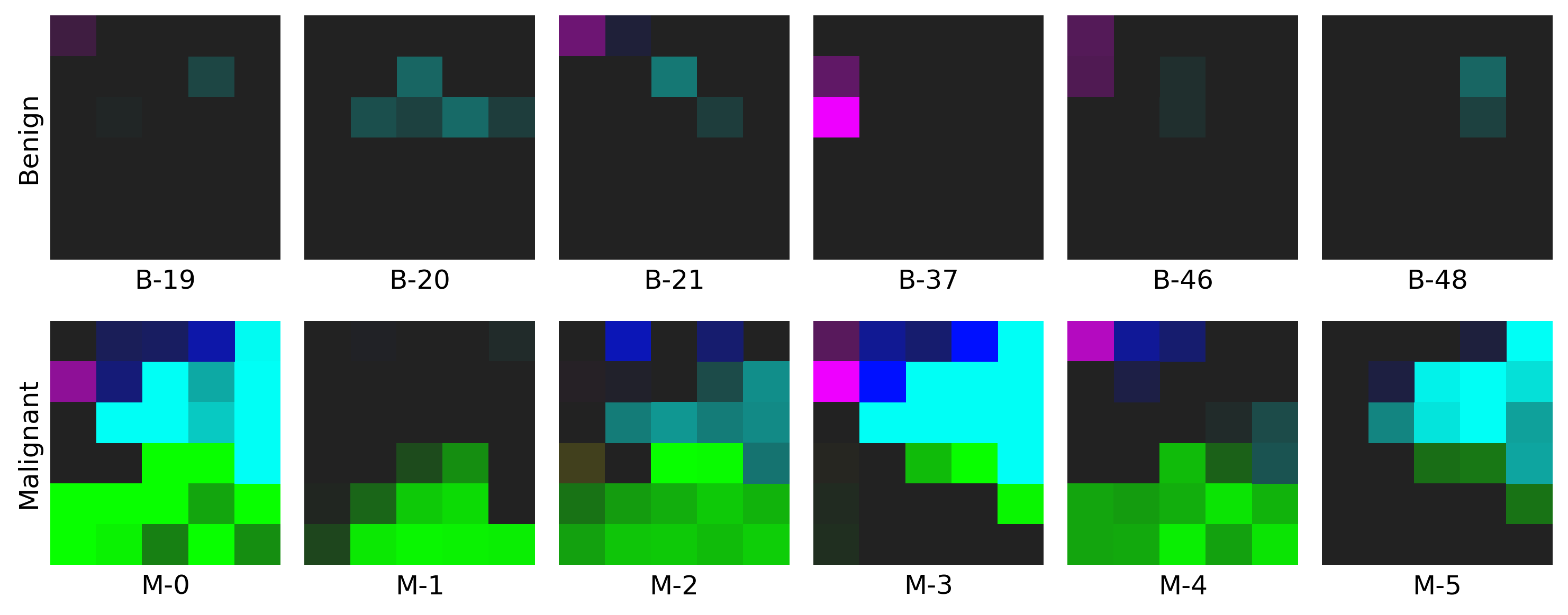
Example for Restructured Fmaps
- The example on WDBC dataset: click here to find out more!
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file aggmap-1.2.0.tar.gz.
File metadata
- Download URL: aggmap-1.2.0.tar.gz
- Upload date:
- Size: 55.2 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.8.17
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
01e3c14845501ee470cb4bfe668db0961f5a05a618a1836d930c54df0a15adae
|
|
| MD5 |
3da2fa8b23d95ee94197adda01d56df7
|
|
| BLAKE2b-256 |
b20ea757eab527f4dc5dd84ef356cf2534568e126bd603ac9e23384abb20bb1b
|
File details
Details for the file aggmap-1.2.0-py3-none-any.whl.
File metadata
- Download URL: aggmap-1.2.0-py3-none-any.whl
- Upload date:
- Size: 58.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.8.17
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
ffe9051353d5f7390186f49417e9a7fa82da4c1f6e18c53515a2d1cd8e8bc64e
|
|
| MD5 |
5a73a3d1a41e1032ffaee13bb3a7daba
|
|
| BLAKE2b-256 |
143d7281347671bd1dafbf12f4f9d9d9f057e737c9957077cde534e63d24b837
|