A console-based sequence alignment viewer
Project description
alv: a command-line alignment viewer
View your DNA or protein multiple-sequence alignments right at your command line. No need to launch a GUI!
Note: alv requires Python v3.6 or later. Earlier versions may also work, but this has not been
tested.
Latest feature additions
-
If you have more than one alignment in your input file, then the first alignment is output unless you use the --alignment-index (-ai) option to choose another.
-
alvis now adapted for use in Python notebooks (tested on Jupyter) through two convenience functions 'view' and 'glimpse'. Both functions take a BioPython alignment object and outputs a view of the alignment.Writing
from Bio import AlignIO msa = AlignIO.read('PF00005.fa', 'fasta') import alv alv.view(msa)in a Jupyter notebook cell and evaluating will yield a colored alignment in the
alvstyle.For large alignments, the glimpse function is convenient since a subset of the alignment, selected as an easily detected conserved region, is shown.
alv.glimpse(msa)Look for more usage information view
help(alv.view)in a notebook cell.
Features
- Command-line based, no GUI, so easy to script viewing of many (typically small) MSAs.
- Reads alignments in FASTA, Clustal, PHYLIP, NEXUS, and Stockholm formats, from file or
stdin. - Output is formatted to suit your terminal. You can also set the alignment width with option
-w. - Can color alignments of coding DNA by codon's translations to amino acids.
- Guesses sequence type (DNA/RNA/AA/coding) by default. You can override with option
-t. - Order sequence explicitly, alphabetically, or by sequence similarity.
- Restrict coloring to where you don't have indels or where there is a lot of conservation.
- Focus on variable columns with the options
--only-variableand--only-variable-excluding-indels, contributed by nikostr, that constrains coloring to columns with variation and variation not counting indels. - The command
alv -g huge_msa.fadisplays cut-out of the MSA, guaranteed to fit one terminal page without scrolling or MSA line breaking, that is supposed to give you an idea of alignment quality and contents. - Write
alv -r 20 huge_msa.fato get a view of the MSA containing only 20 randomly selected sequences.
Install
Recommended installation is:
pip install --upgrade pip
pip install alv
If you have a half-modern BioPython installed, Python v3.4 should work.
BioPython is a dependency and will only get installed automatically with pip install alv
if you are using Python v3.6 or later, because BioPython was apparently not on PyPi before that.
Install the commandline tool system-wide
If you would like to install the alv commandline tool system-wide, so that it
is accessible regardless of which python environment you have loaded, or if you
have loaded one at all, one way to do this is using the
pipx installer.
After installing pipx using it's official installation
instructions you
can install alv with:
pipx install alv
After this, alv should be available as a command regardless of Python
environment.
(Note that this only works for installing the alv commandline tool, not the
Python library!)
Examples
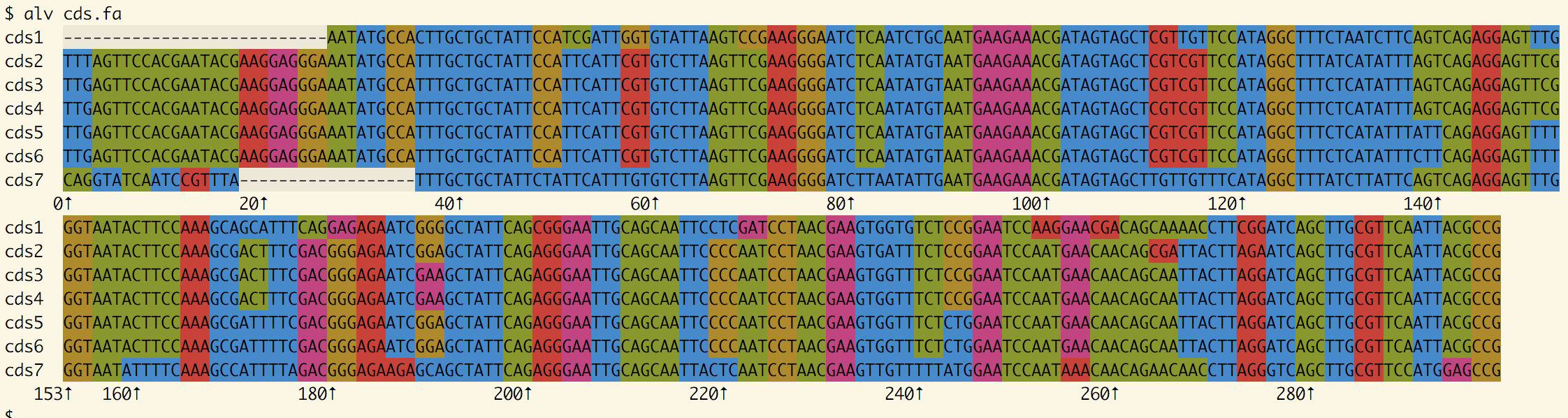
Quick viewing of a small alignment:
alv msa.fa
This autodetects sequence type (AA, DNA, RNA, coding DNA), colors the sequences, and formats the
alignment for easy viewing in your terminal.
When applying alv to an alignment of coding DNA, the coding property is autodetected and colors are therefore applied to codons instead
of nucleotides.
View three sequences, accessions a, b, and c, from an alignment:
alv -so a,b,c msa.fa
Feed alignment to less, for paging support.
alv -k msa.fa | less -R
The -k option ensures that alv keeps coloring the alignment (by default, piping
and redirection removes colors), and the -R option instructs less to interpret color codes.
Choose to view a sub-alignment:
alv -sa 30 60 msa.fa
This selects and views columns 30 to 59 of msa.fa, keeping track of the "original" columns indexes in the output.
For developers
- Run
pip install -e .to get an "editable" install, while coding. - Run
python -m buildto prepare a distributable file. - Run
python -m unittest discoverto run unit tests.
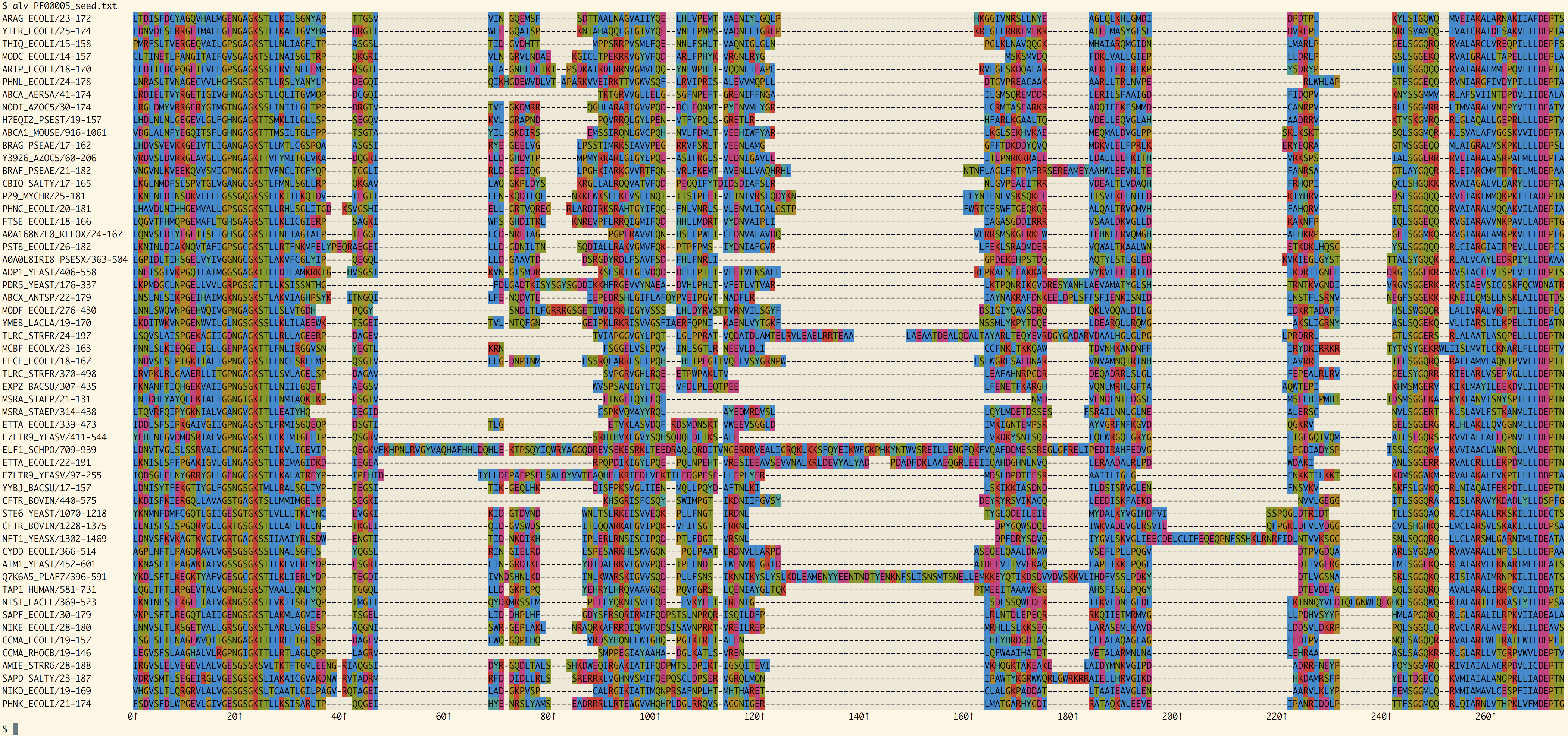
Screenshots
Full PFAM domain
All of the sequences in PFAM's seed alignment for PF00005
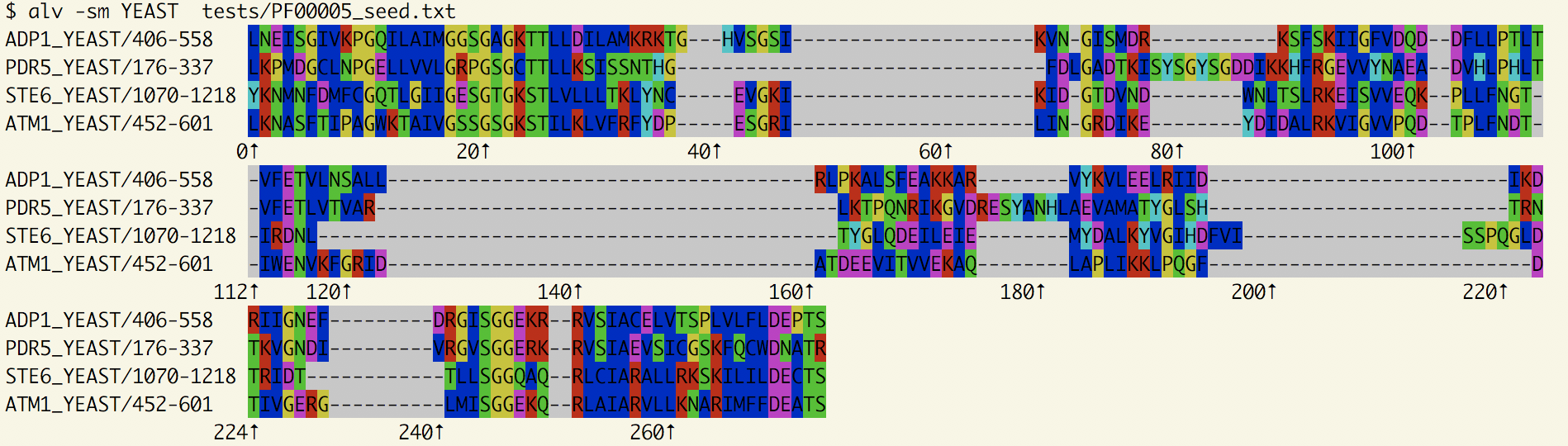
Yeast sequences from PF00005
Using the option -sm YEAST, we reduce the alignment to the ones with a matching accession.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file alv-1.8.1.tar.gz.
File metadata
- Download URL: alv-1.8.1.tar.gz
- Upload date:
- Size: 36.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.10.19
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
1ef20c45b99c932f4325fd9ed82d163091181bc09da852755f5fb87f1d04b721
|
|
| MD5 |
36673ad36c926ec4a16fc4c6969eb4c9
|
|
| BLAKE2b-256 |
349efe097ab4e9b28bf1e27e4dd41a4a126e92fa7f52faf129dad26f673dd8fb
|
File details
Details for the file alv-1.8.1-py3-none-any.whl.
File metadata
- Download URL: alv-1.8.1-py3-none-any.whl
- Upload date:
- Size: 33.0 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.10.19
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
2547ad665ea591d5c4bec25cfdc98d980ebb6e290adfb7dfc23bb557a0364f11
|
|
| MD5 |
0384f449007895ea9158b310b039b895
|
|
| BLAKE2b-256 |
38d5209228428247531efb10e9897ed8a8d1d18846953fa7f073a6028a470712
|