Bam2Tensor
Project description
bam2tensor
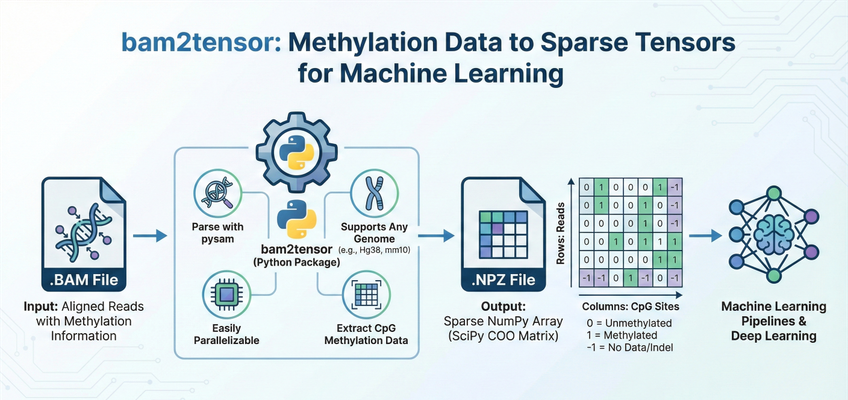
bam2tensor is a Python package for converting bisulfite-sequencing .bam files to sparse tensor representations of DNA methylation data. It extracts read-level methylation states from CpG sites and outputs efficient sparse COO matrices as .npz files, ready for deep learning pipelines.
Table of Contents
- Features
- Requirements
- Installation
- Quick Start
- Usage
- Output Data Structure
- Supported Aligners
- Performance Tips
- API Reference
- Contributing
- License
- Credits
Features
- BAM Parsing: Efficiently parses
.bamfiles using pysam - Complete CpG Extraction: Extracts methylation data from all CpG sites genome-wide
- Multi-Genome Support: Works with any reference genome (GRCh38/hg38, T2T-CHM13, mm10, etc.)
- Sparse Storage: Stores data in sparse COO matrix format for memory-efficient loading
- NumPy/SciPy Integration: Exports to
.npzfiles compatible with NumPy and SciPy - Efficient Algorithm: Linear-scan algorithm ensures minimal memory usage with no read duplication
- Batch Processing: Process multiple BAM files with directory recursion
- Caching: CpG site indexing is cached to accelerate repeated runs on the same genome
- Quality Filtering: Configurable mapping quality thresholds
Requirements
- Python 3.10 or higher
- A reference genome FASTA file (must match the genome used for alignment)
- Indexed BAM files (
.bamwith corresponding.bam.baiindex files)
Dependencies
Core dependencies are automatically installed:
pysam- BAM file handlingbiopython- FASTA parsingscipy- Sparse matrix operationsnumpy- Numerical operationsclick- Command-line interfacetqdm- Progress bars
Installation
From PyPI (Recommended)
pip install bam2tensor
From Source
git clone https://github.com/mcwdsi/bam2tensor.git
cd bam2tensor
pip install .
Development Installation
git clone https://github.com/mcwdsi/bam2tensor.git
cd bam2tensor
pip install poetry
poetry install
Quick Start
# Basic usage with a single BAM file
bam2tensor \
--input-path sample.bam \
--reference-fasta GRCh38.fa \
--genome-name hg38
# This creates: sample.methylation.npz
Usage
Basic Usage
Process a single bisulfite-sequencing BAM file:
bam2tensor \
--input-path /path/to/aligned_reads.bam \
--reference-fasta /path/to/reference.fa \
--genome-name hg38
This will:
- Parse the reference FASTA to identify all CpG sites (cached for future runs)
- Extract methylation states from each read in the BAM file
- Output a sparse matrix to
aligned_reads.methylation.npz
Processing Multiple BAM Files
Process all BAM files in a directory recursively:
bam2tensor \
--input-path /path/to/bam_directory/ \
--reference-fasta /path/to/reference.fa \
--genome-name hg38 \
--verbose
Each BAM file will generate a corresponding .methylation.npz file in the same location.
Using a Custom Genome
For non-human genomes or custom chromosome sets:
# Mouse genome (mm10)
bam2tensor \
--input-path mouse_sample.bam \
--reference-fasta mm10.fa \
--genome-name mm10 \
--expected-chromosomes "chr1,chr2,chr3,chr4,chr5,chr6,chr7,chr8,chr9,chr10,chr11,chr12,chr13,chr14,chr15,chr16,chr17,chr18,chr19,chrX,chrY"
# T2T-CHM13 human genome
bam2tensor \
--input-path sample.bam \
--reference-fasta chm13v2.0.fa \
--genome-name T2T-CHM13 \
--expected-chromosomes "chr1,chr2,chr3,chr4,chr5,chr6,chr7,chr8,chr9,chr10,chr11,chr12,chr13,chr14,chr15,chr16,chr17,chr18,chr19,chr20,chr21,chr22,chrX,chrY"
Command-Line Options
Usage: bam2tensor [OPTIONS]
Extract read-level methylation data from an aligned .bam file and export
the data as a SciPy sparse matrix.
Options:
--version Show the version and exit.
--input-path PATH Input .bam file OR directory to recursively
process. [required]
--genome-name TEXT A custom string referring to your genome
name, used to save a cache file (e.g. hg38,
hg38-no-alt, etc.). [required]
--expected-chromosomes TEXT A comma-separated list of chromosomes to
expect in the .fa genome. Defaults to hg38
chromosomes (chr1-chr22, chrX, chrY).
--reference-fasta PATH Reference genome FASTA file (critical to
determine CpG sites). [required]
--quality-limit INTEGER Quality filter for aligned reads (default =
20).
--verbose Verbose output.
--skip-cache De-novo generate CpG sites (slow).
--debug Debug mode (extensive validity checking +
debug messages).
--overwrite Overwrite output file if it exists.
--help Show this message and exit.
Option Details
| Option | Description |
|---|---|
--input-path |
Path to a single .bam file or a directory. If a directory is provided, all .bam files are processed recursively. |
--genome-name |
An identifier for your reference genome (e.g., hg38, mm10). Used to name the cache file for CpG site positions. |
--expected-chromosomes |
Comma-separated list of chromosome names to process. Chromosomes not in this list are skipped. Defaults to human autosomes + sex chromosomes. |
--reference-fasta |
Path to the reference genome FASTA file. Must match the genome used for alignment. |
--quality-limit |
Minimum mapping quality score (MAPQ) for reads to be included. Default is 20. |
--verbose |
Enable detailed progress output including per-chromosome progress bars. |
--skip-cache |
Force regeneration of CpG site cache. Useful if you've modified the reference or chromosome list. |
--debug |
Enable extensive validation and debug output. Slower but useful for troubleshooting. |
--overwrite |
Overwrite existing .methylation.npz files. Without this flag, existing outputs are skipped. |
Output Data Structure
bam2tensor generates one .npz file per input BAM file. Each file contains a SciPy sparse COO matrix with the following structure:
| Dimension | Represents |
|---|---|
| Rows | Unique reads (primary alignments that pass quality filters) |
| Columns | CpG sites (ordered by genomic position across all chromosomes) |
Methylation State Values
| Value | Meaning |
|---|---|
1 |
Methylated (cytosine preserved as C) |
0 |
Unmethylated (cytosine converted to T by bisulfite treatment) |
-1 |
No data (indel, SNV, or site not covered by read) |
Note: Sparse matrices only store non-zero values. Positions with value 0 (unmethylated) are stored, but positions not covered by a read are simply absent from the matrix.
Loading Output Files
import scipy.sparse
import numpy as np
# Load the sparse matrix
methylation_matrix = scipy.sparse.load_npz("sample.methylation.npz")
print(f"Matrix shape: {methylation_matrix.shape}")
print(f"Number of reads: {methylation_matrix.shape[0]}")
print(f"Number of CpG sites: {methylation_matrix.shape[1]}")
print(f"Non-zero entries: {methylation_matrix.nnz}")
print(f"Sparsity: {1 - methylation_matrix.nnz / np.prod(methylation_matrix.shape):.4%}")
Converting to Dense Arrays
For small regions or when dense operations are needed:
# Convert entire matrix to dense (warning: may use significant memory)
dense_matrix = methylation_matrix.toarray()
# Convert to CSR format for efficient row slicing
csr_matrix = methylation_matrix.tocsr()
# Get methylation data for reads 0-99
subset = csr_matrix[0:100, :].toarray()
# Convert to CSC format for efficient column slicing
csc_matrix = methylation_matrix.tocsc()
# Get data for CpG sites 1000-1099
cpg_subset = csc_matrix[:, 1000:1100].toarray()
Working with Genomic Coordinates
To map between matrix column indices and genomic coordinates, use the GenomeMethylationEmbedding class:
from bam2tensor.embedding import GenomeMethylationEmbedding
# Load or recreate the embedding used during extraction
embedding = GenomeMethylationEmbedding(
genome_name="hg38",
expected_chromosomes=["chr" + str(i) for i in range(1, 23)] + ["chrX", "chrY"],
fasta_source="/path/to/GRCh38.fa",
)
# Convert matrix column index to genomic position
chrom, pos = embedding.embedding_to_genomic_position(12345)
print(f"Column 12345 corresponds to {chrom}:{pos}")
# Convert genomic position to matrix column index
col_idx = embedding.genomic_position_to_embedding("chr1", 10525)
print(f"chr1:10525 is at column {col_idx}")
# Get total number of CpG sites
print(f"Total CpG sites: {embedding.total_cpg_sites:,}")
Example: Analyzing Methylation Patterns
import scipy.sparse
import numpy as np
# Load the data
matrix = scipy.sparse.load_npz("sample.methylation.npz")
csr = matrix.tocsr()
# Calculate per-CpG methylation rates (excluding -1 values)
methylation_rates = []
for cpg_idx in range(matrix.shape[1]):
col_data = csr.getcol(cpg_idx).toarray().flatten()
# Filter out -1 (no data) and positions with no coverage
valid_data = col_data[(col_data >= 0)]
if len(valid_data) > 0:
rate = np.mean(valid_data)
else:
rate = np.nan
methylation_rates.append(rate)
methylation_rates = np.array(methylation_rates)
print(f"Mean methylation rate: {np.nanmean(methylation_rates):.2%}")
print(f"CpG sites with coverage: {np.sum(~np.isnan(methylation_rates)):,}")
Example: Integration with PyTorch
import torch
import scipy.sparse
import numpy as np
# Load sparse matrix
matrix = scipy.sparse.load_npz("sample.methylation.npz")
# Convert to PyTorch sparse tensor
coo = matrix.tocoo()
indices = torch.LongTensor(np.vstack((coo.row, coo.col)))
values = torch.FloatTensor(coo.data)
shape = torch.Size(coo.shape)
sparse_tensor = torch.sparse_coo_tensor(indices, values, shape)
print(f"PyTorch sparse tensor shape: {sparse_tensor.shape}")
# For models that need dense input (specific region)
region_start, region_end = 0, 1000
dense_region = matrix.tocsc()[:, region_start:region_end].toarray()
dense_tensor = torch.FloatTensor(dense_region)
Supported Aligners
bam2tensor supports BAM files from bisulfite-aware aligners that include strand information tags:
| Aligner | Tag | Values |
|---|---|---|
| Biscuit | YD |
f (forward/OT/CTOT), r (reverse/OB/CTOB) |
| gem3 / Blueprint | XB |
C (forward), G (reverse) |
These tags indicate which strand the original bisulfite-converted DNA came from, which is essential for correctly interpreting C/T as methylated/unmethylated.
Performance Tips
-
Use the cache: The first run on a new genome builds a CpG site index, which is cached. Subsequent runs are much faster.
-
Process in parallel: bam2tensor processes one BAM at a time, but you can run multiple instances in parallel on different BAM files:
# Using GNU parallel find /data/bams -name "*.bam" | parallel -j 4 \ bam2tensor --input-path {} --reference-fasta ref.fa --genome-name hg38
-
Ensure BAM files are indexed: Each BAM file requires a corresponding
.bam.baiindex file. Create with:samtools index sample.bam
-
Use SSDs: Both reading BAM files and writing output benefit from fast storage.
-
Memory considerations: Memory usage scales with the number of CpG sites (columns) rather than reads. For human genomes (~28M CpG sites), expect moderate memory usage.
API Reference
bam2tensor.embedding.GenomeMethylationEmbedding
Main class for managing CpG site positions and coordinate conversions.
GenomeMethylationEmbedding(
genome_name: str, # Identifier for caching
expected_chromosomes: list, # List of chromosome names to process
fasta_source: str, # Path to reference FASTA
skip_cache: bool = False, # Force regeneration of cache
verbose: bool = False # Enable verbose output
)
Key Methods:
embedding_to_genomic_position(embedding: int) -> tuple[str, int]- Convert column index to (chromosome, position)genomic_position_to_embedding(chrom: str, pos: int) -> int- Convert genomic position to column index
Key Attributes:
total_cpg_sites: int- Total number of CpG sites across all chromosomescpg_sites_dict: dict[str, list[int]]- Dictionary mapping chromosome names to lists of CpG positions
bam2tensor.functions.extract_methylation_data_from_bam
Core function for extracting methylation data from a BAM file.
extract_methylation_data_from_bam(
input_bam: str, # Path to BAM file
genome_methylation_embedding: GenomeMethylationEmbedding, # Embedding object
quality_limit: int = 20, # Minimum MAPQ
verbose: bool = False, # Enable verbose output
debug: bool = False # Enable debug output
) -> scipy.sparse.coo_matrix
Returns: A SciPy COO sparse matrix with shape (n_reads, n_cpg_sites).
Contributing
Contributions are welcome! Please see the Contributor Guide for guidelines on:
- Setting up a development environment
- Running tests
- Code style requirements
- Submitting pull requests
Development Commands
# Install development dependencies
poetry install
# Run all checks (linting, type checking, tests)
nox
# Run specific checks
nox --session=tests # Run pytest
nox --session=mypy # Type checking
nox --session=pre-commit # Linting
# Format code
poetry run black src tests
poetry run ruff check --fix src tests
License
Distributed under the terms of the MIT license, bam2tensor is free and open source software.
Issues
If you encounter any problems, please file an issue with:
- A description of the problem
- Steps to reproduce
- Your Python version and operating system
- Relevant error messages or logs
Credits
This project is developed and maintained by Nick Semenkovich (@semenko), as part of the Medical College of Wisconsin's Data Science Institute.
This project was generated from Statistics Norway's SSB PyPI Template.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file bam2tensor-1.5.tar.gz.
File metadata
- Download URL: bam2tensor-1.5.tar.gz
- Upload date:
- Size: 25.9 kB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
9922ace77da92ca1fe8f3ebddc9a4211aac82e07da42e005a253c1b165e086b1
|
|
| MD5 |
fa5ec93149eecc9e56b7dcff64294d40
|
|
| BLAKE2b-256 |
6d0347925fa16d4f9830c8c1f55ef68eedce0d73c62c47d7a787de2293067563
|
Provenance
The following attestation bundles were made for bam2tensor-1.5.tar.gz:
Publisher:
release.yml on mcwdsi/bam2tensor
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
bam2tensor-1.5.tar.gz -
Subject digest:
9922ace77da92ca1fe8f3ebddc9a4211aac82e07da42e005a253c1b165e086b1 - Sigstore transparency entry: 802542896
- Sigstore integration time:
-
Permalink:
mcwdsi/bam2tensor@e27f867f8f9b3af52b88b9876d73aa5ccb3a225c -
Branch / Tag:
refs/heads/main - Owner: https://github.com/mcwdsi
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
release.yml@e27f867f8f9b3af52b88b9876d73aa5ccb3a225c -
Trigger Event:
push
-
Statement type:
File details
Details for the file bam2tensor-1.5-py3-none-any.whl.
File metadata
- Download URL: bam2tensor-1.5-py3-none-any.whl
- Upload date:
- Size: 23.6 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
c1e2dab4332a13c84c757232abb70f1c2098c88b1c10914fe89dbd8893568cb4
|
|
| MD5 |
4defecb76e2d0f4e91f36d459ce90d27
|
|
| BLAKE2b-256 |
d3d379d2ea8fd2bcfd7c2288657087d3dd4abc5b43777e85326ea2feb793768c
|
Provenance
The following attestation bundles were made for bam2tensor-1.5-py3-none-any.whl:
Publisher:
release.yml on mcwdsi/bam2tensor
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
bam2tensor-1.5-py3-none-any.whl -
Subject digest:
c1e2dab4332a13c84c757232abb70f1c2098c88b1c10914fe89dbd8893568cb4 - Sigstore transparency entry: 802542985
- Sigstore integration time:
-
Permalink:
mcwdsi/bam2tensor@e27f867f8f9b3af52b88b9876d73aa5ccb3a225c -
Branch / Tag:
refs/heads/main - Owner: https://github.com/mcwdsi
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
release.yml@e27f867f8f9b3af52b88b9876d73aa5ccb3a225c -
Trigger Event:
push
-
Statement type: