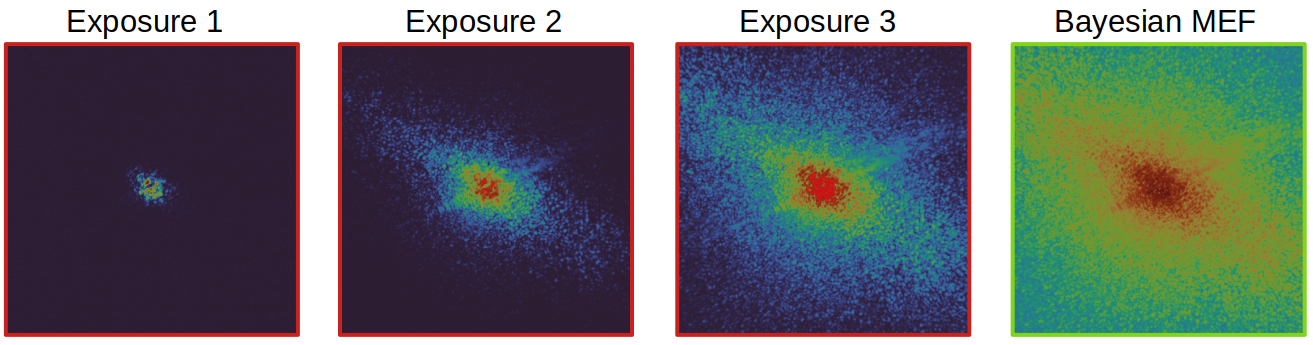
Python implementation of the Bayesian MEF method
Project description
Bayesian MEF
Bayesian multi-exposure image fusion (MEF) is a general-purpose algorithm to achieve robust high dynamic range (HDR) imaging, particularly in scenarios with low signal-to-noise ratio (SNR) or variations in illumination intensity. This approach could especially be useful for high quality phase retrieval in coherent diffractive imaging (CDI). The algorithm, detailed in this publication "Bayesian multi-exposure image fusion for robust high dynamic range ptychography", explains the method and demonstrates the benefits for ptychography. To reproduce the results in the paper, see this section. However, to get started, please install the package and check the demo usage below.
To install this package from PyPI,
pip install bayes_mef
Usage
A minimal example demonstrating the usage of BayesianMEF by simulating some data.
from bayes_mef import BayesianMEF
from skimage.data import camera
import numpy as np
# simulation params
truth = camera()
background = 60 # some background
times = np.array([0.1, 1, 10]) # exposure times or equivalently flux factors
threshold = 1500 # detector limit
# poisson data based on image formation model that is overexposed
data = [np.random.poisson(time * truth + background) for time in times]
data_saturated = np.clip(data, None, threshold, dtype="float")
# Bayesian MEF with optional field `update_fluxes`. Set it to `True` when
# flux factors (exposure times) are not accurately known.
mef_em = BayesianMEF(data_saturated, threshold, times, background, update_fluxes=False)
mef_em.run(n_iter=100)
fused_em = mef_em.fused_image.copy()
Additionally, one can also use the ConventionalMEF method as given in the paper.
from bayes_mef import ConventionalMEF
mef_mle = ConventionalMEF(data_saturated, threshold, times, background)
mef_mle.mle()
fused_mle = mef_mle.fused_image.copy()
Parallelized implementatation
In ptychography, one needs to fuse diffraction patterns for every scan position. Processing this for data over all scan positions can be slow for an iterative algorithm. Therefore, one can use the parallelized implementation for MEF LaunchMEF. Check the example below
from bayes_mef import LaunchMEF
launch_mef = LaunchMEF(
ptychogram_stack, # ptychogram shape (n_exposures, n_scans, dp_x, dp_y)
background, # background shape (n_exposures, n_scans, dp_x, dp_y)
flux_factors=None, # if set to `None`, calculates automatically
threshold=None, # if set to `None`, calculates automatically
update_fluxes=False, # set to `True` if you want to update fluxes
)
# runs Bayesian MEF in parallel by defining the number of CPUs `n_cpus`;
# returns the fused diffraction patterns with shape (n_scans, dp_x, dp_y) and updated flux factors
n_cpus = 20
n_iter = 150
fused_ptyem_stack, em_flux_factors = launch_mef.run_em(n_iter, n_cpus)
For a detailed usage, please check synthetic_mef.py that uses synthetic ptychography data.
Reproducing results
To reproduce the ptychographic reconstruction results from the paper, please install the package in development mode and download the necessary data. This is explained in the following steps:
- Please clone this repository and create a conda environment.
git clone https://github.com/microscopic-image-analysis/bayes-mef.git cd bayes-mef conda create --name bayes-mef-venv python=3.11.5
- Now activate the environment and install the pinned dependencies.
conda activate bayes-mef-venv pip install -r requirements.txt
[!NOTE] The package is maintained via python package manager Poetry, so instead of using
pipfor installing the package and its requirements, installation can also simply be done aspoetry install.
-
Download the data from Zenodo with the following command:
./download_data.sh
-
Optional: For faster ptychographic reconstructions using GPU, please install
cupyas given under its installation guide. -
Run files from the scripts/ directory for plotting the results.
Citation
If you found this algorithm or the publication useful, please cite us at:
@article{Kodgirwar:24,
author = {Shantanu Kodgirwar and Lars Loetgering and Chang Liu and Aleena Joseph and Leona Licht and Daniel S. Penagos Molina and Wilhelm Eschen and Jan Rothhardt and Michael Habeck},
journal = {Opt. Express},
number = {16},
pages = {28090--28099},
publisher = {Optica Publishing Group},
title = {Bayesian multi-exposure image fusion for robust high dynamic range ptychography},
volume = {32},
month = {Jul},
year = {2024},
url = {https://opg.optica.org/oe/abstract.cfm?URI=oe-32-16-28090},
doi = {10.1364/OE.524284},
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file bayes_mef-0.1.9.tar.gz.
File metadata
- Download URL: bayes_mef-0.1.9.tar.gz
- Upload date:
- Size: 13.2 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/2.1.3 CPython/3.13.5 Darwin/24.6.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
c998a199b584ec0f31e71fdedccd32e963e1b2c68c9393256dd3be909b4868fd
|
|
| MD5 |
f5caa43705ae4fd48e2c8d2a4d4149aa
|
|
| BLAKE2b-256 |
e1e351fb24827b8608eddbd1d206aa634277d351d45b88ea7e922dbf9dec1935
|
File details
Details for the file bayes_mef-0.1.9-py3-none-any.whl.
File metadata
- Download URL: bayes_mef-0.1.9-py3-none-any.whl
- Upload date:
- Size: 12.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/2.1.3 CPython/3.13.5 Darwin/24.6.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
a2a9f49391599b2390e10d8ac5b792d34b1b903492428027505d3dffbde4a8e1
|
|
| MD5 |
d40df5c27500b3ac22fbbacd0b0a8fb5
|
|
| BLAKE2b-256 |
030f61bb522d49af1c209daaa9c5021a39e03aeaf96d4f88a3d5d29ca41dd66c
|