Bio Matrix Topology (BiMaTo) is a library containing all the biopolymer matrix topology analyses published by the Biological Physics Group, (BIP), Peter Debye Institute, University Leipzig, Germany.
Project description
Bio Matrix Topology (BiMaTo) is a library containing all the biopolymer matrix topology analyses published by the Biological Physics Group, (BIP), Peter Debye Institute, University Leipzig, Germany.
Documentation can be found here.
How to install
bimato uses Python3.8 and up. Installation is trivial:
pip install bimato
Exemplary analysis workflow
This is an exemplary workflow to analyze pore sizes of two different collagen scaffolds. The matrices have been fluorescently stained and 3D images were recorded using an LSM.
Usually, we have for example different collagen scaffolds and want to compare structural parameters. For this, we would load several images, calculate their structural parameters and plot them. Below is an exemplary workflow for this:
load each image in the LIF file
analyze it
extract meta-data such as collagen concentration from image name
concatenate this data to global DataFrame
plot comparison boxplot
Pore-Size
We load a lif file with multiple samples per collagen concentration and analyze these in a loop:
import pandas as pd
from readlif.reader import LifFile
import seaborn as sns
import bimato
lif_file = LifFile("/path/to/sample.lif")
df_poresize = list()
for lif_image in lif_file.get_iter_image():
data = bimato.utils.read_lif_image(lif_image)
bw = bimato.get_binary(data)
sampling = {
'x': 1/lif_image.info["scale"][0],
'y': 1/lif_image.info["scale"][1],
'z': 1/lif_image.info["scale"][2]
}
df_tmp = bimato.get_pore_sizes(binary=bw, sampling=sampling)
df_tmp['Concentration [g/l]'] = lif_image.name
df_poresize.append(df_tmp)
df_poresize = pd.concat(df_poresize)
g = sns.catplot(
data=df_poresize,
kind='box',
x='Concentration [g/l]',
y='Diameter [µm],
)
g.set_ylabels("Pore-size [µm]")Resulting in the following plot:
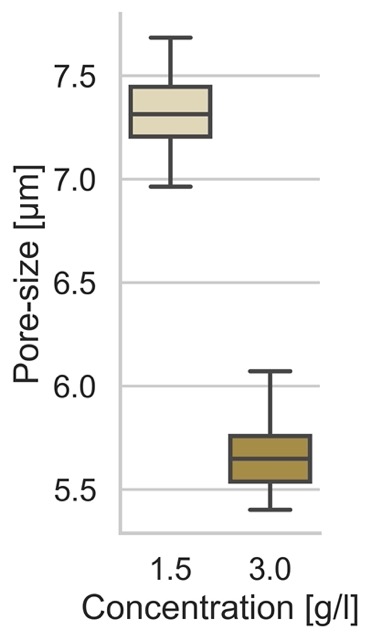
Inhomogeneity
We load a lif file with multiple samples per collagen concentration and analyze these in a loop:
import pandas as pd
from readlif.reader import LifFile
import seaborn as sns
import bimato
lif_file = LifFile("/path/to/sample.lif")
df_inhomogeneity = list()
for lif_image in lif_file.get_iter_image():
data = bimato.utils.read_lif_image(lif_image)
bw = bimato.get_binary(data)
sampling = {
'x': 1/lif_image.info["scale"][0],
'y': 1/lif_image.info["scale"][1],
'z': 1/lif_image.info["scale"][2]
}
df_tmp = bimato.poresize.get_fragmented_poresizes(binary=bw, sampling=sampling, part_size_micron=30)
df_tmp['Inhomogeneity'] = bimato.poresize.calc_inhomogeneity(df_tmp)
df_tmp['Concentration [g/l]'] = lif_image.name
df_inhomogeneity.append(df_tmp)
df_inhomogeneity = pd.concat(df_inhomogeneity)
g = sns.catplot(
data=df_poresize,
kind='box',
x='Concentration [g/l]',
y='Inhomogeneity,
)Resulting in the following plot:
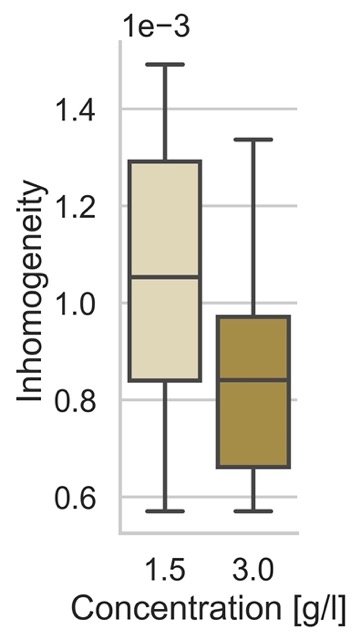
How to cite
Fischer T, Hayn A, Mierke CT (2019) Fast and reliable advanced two-step pore-size analysis of biomimetic 3D extracellular matrix scaffolds. Scientific Reports 9:8352. https://doi.org/10.1038/s41598-019-44764-5
Hayn A, Fischer T, Mierke CT (2020) Inhomogeneities in 3D Collagen Matrices Impact Matrix Mechanics and Cancer Cell Migration. Front Cell Dev Biol 8:593879. https://doi.org/10.3389/fcell.2020.593879
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file bimato-2022.2.1.tar.gz.
File metadata
- Download URL: bimato-2022.2.1.tar.gz
- Upload date:
- Size: 25.4 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.2.2 CPython/3.10.8 Darwin/21.6.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
b3d1439493761c7672bf277bc656c7d4a7823e6bf948232c9d5fe43ed1daa8e2
|
|
| MD5 |
909c50a84536e48e025cd17864592001
|
|
| BLAKE2b-256 |
d515b40675546881943429617f48ac4d134f19aebc7db5d67a85bd91d13c1153
|
File details
Details for the file bimato-2022.2.1-py3-none-any.whl.
File metadata
- Download URL: bimato-2022.2.1-py3-none-any.whl
- Upload date:
- Size: 28.5 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.2.2 CPython/3.10.8 Darwin/21.6.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
20d288fa471b915486761569e47affe44fd4f4e399de8970436c7e81328cd020
|
|
| MD5 |
4c5486e68c6f06e4a1b164e12b2d59c8
|
|
| BLAKE2b-256 |
dd305d228c2f86ea497879fa9cdab7fe700524f23be3f1e9345b6162b1de347b
|
















