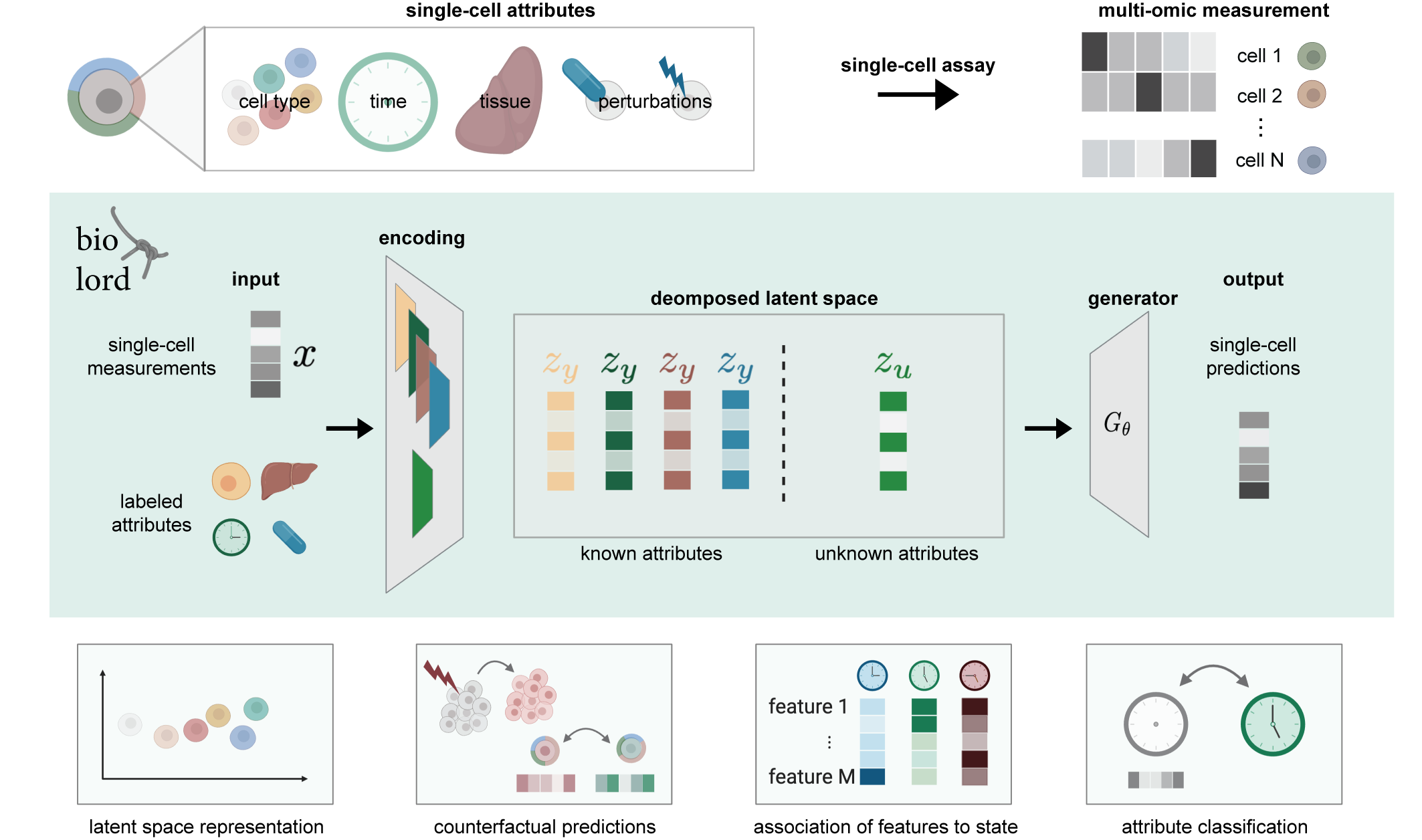
A deep generative framework for disentangling known and unknown attributes in single-cell data.
Project description
biolord - biological representation disentanglement
A deep generative framework for disentangling known and unknown attributes in single-cell data.
We assume partial supervision over known attributes (categorical or ordered) along with single-cell measurements. Given the partial supervision biolord finds a decomposed latent space, and provides a generative model to obtain single-cell measurements for different cell states.
For more details read our pubication in Nature Biotechnology.
Getting started
Please refer to the documentation.
Installation
There are several alternative options to install biolord:
-
Install the latest release of biolord from PyPI:
pip install biolord
-
Install the latest development version:
pip install git+https://github.com/nitzanlab/biolord.git@main
Release notes
See the changelog.
Contact
Feel free to contact us by mail. If you found a bug, please use the issue tracker.
Citation
@article{piran2024disentanglement,
title={Disentanglement of single-cell data with biolord},
author={Piran, Zoe and Cohen, Niv and Hoshen, Yedid and Nitzan, Mor},
journal={Nature Biotechnology},
pages={1--6},
year={2024},
publisher={Nature Publishing Group US New York}
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file biolord-0.0.3.tar.gz.
File metadata
- Download URL: biolord-0.0.3.tar.gz
- Upload date:
- Size: 1.6 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.0.0 CPython/3.9.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
67ca596b11138545027e35d562d242c137988597f9041300c11a870c46a33bb2
|
|
| MD5 |
0f9635b83155bacfc40ddf7f192d8e84
|
|
| BLAKE2b-256 |
38a5056ab62582ca5b7b05dfa672934be26dba7a73af9a55fa52096bdb291b51
|
File details
Details for the file biolord-0.0.3-py3-none-any.whl.
File metadata
- Download URL: biolord-0.0.3-py3-none-any.whl
- Upload date:
- Size: 23.1 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.0.0 CPython/3.9.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
f1e9c607332ee2200dad669cf2768ba92b3e1b19121982d915ab91ee1bd5fbe8
|
|
| MD5 |
55944b651756711c7293bb00224b35e9
|
|
| BLAKE2b-256 |
16f212552762c931ae2d744649bc75e0250d7b05f5a34c6b0af17bc5f943e0c5
|