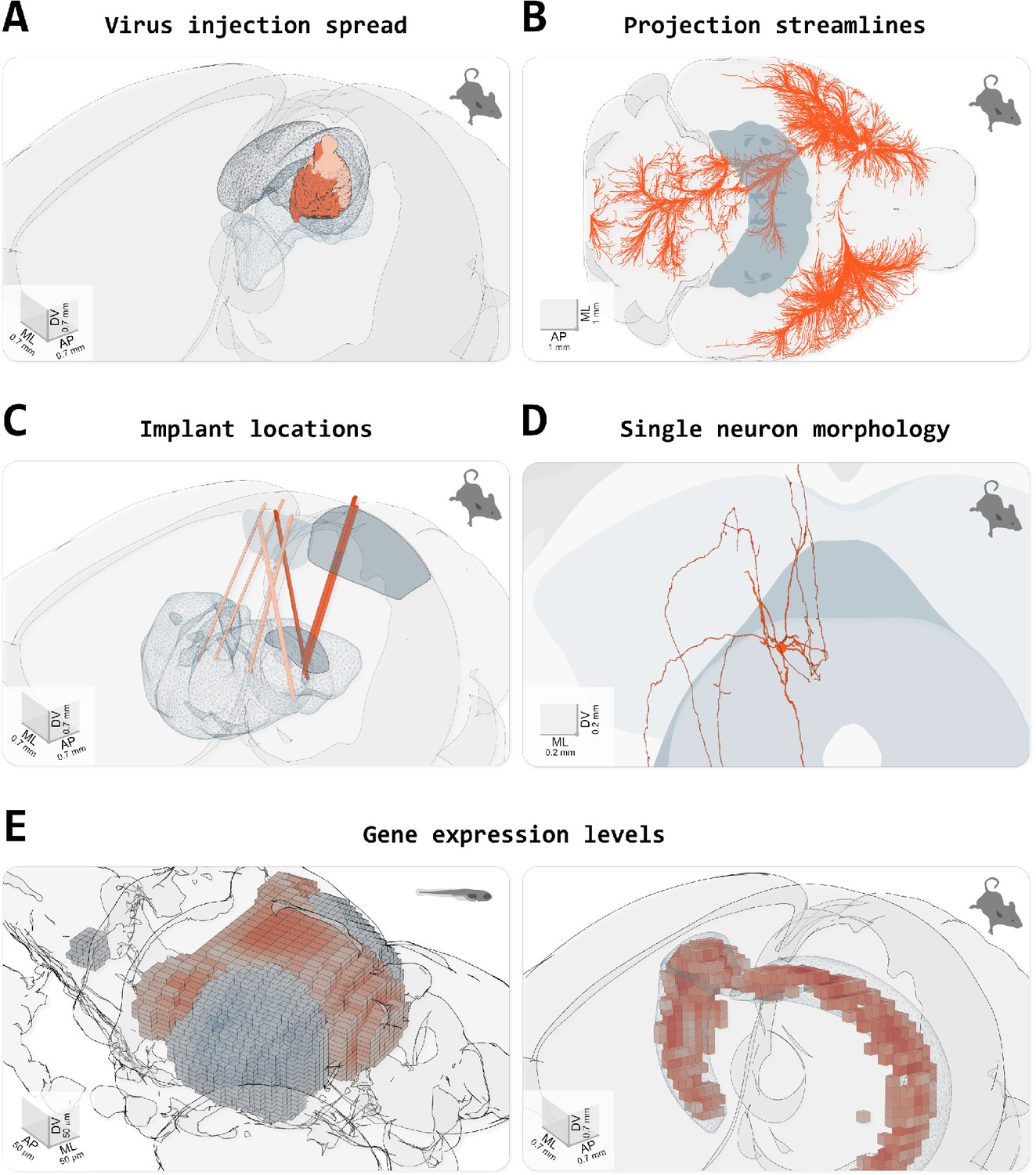
Visualisation and exploration of brain atlases and other anatomical data
Project description
brainrender
A user-friendly python library to create high-quality, 3D neuro-anatomical renderings combining data from publicly available brain atlases with user-generated experimental data.
From: Claudi et al. (2021) Visualizing anatomically registered data with brainrender. eLife
Documentation
brainrender is a project of the BrainGlobe Initiative, which is a collaborative effort to develop a suite of Python-based software tools for computational neuroanatomy. A comprehensive online documentation for brainrender can be found on the BrainGlobe website here.
Furthermore, an open-access journal article describing BrainRender has been published in eLife, available here.
Installation
From PyPI:
pip install brainrender
Quickstart
import random
import numpy as np
from brainrender import Scene
from brainrender.actors import Points
def get_n_random_points_in_region(region, N):
"""
Gets N random points inside (or on the surface) of a mesh
"""
region_bounds = region.mesh.bounds()
X = np.random.randint(region_bounds[0], region_bounds[1], size=10000)
Y = np.random.randint(region_bounds[2], region_bounds[3], size=10000)
Z = np.random.randint(region_bounds[4], region_bounds[5], size=10000)
pts = [[x, y, z] for x, y, z in zip(X, Y, Z)]
ipts = region.mesh.inside_points(pts).coordinates
return np.vstack(random.choices(ipts, k=N))
# Display the Allen Brain mouse atlas.
scene = Scene(atlas_name="allen_mouse_25um", title="Cells in primary visual cortex")
# Display a brain region
primary_visual = scene.add_brain_region("VISp", alpha=0.2)
# Get a numpy array with (fake) coordinates of some labelled cells
coordinates = get_n_random_points_in_region(primary_visual, 2000)
# Create a Points actor
cells = Points(coordinates)
# Add to scene
scene.add(cells)
# Add label to the brain region
scene.add_label(primary_visual, "Primary visual cortex")
# Display the figure.
scene.render()
Seeking help or contributing
We are always happy to help users of our tools, and welcome any contributions. If you would like to get in contact with us for any reason, please see the contact page of our website.
Citing brainrender
If you use brainrender in your scientific work, please cite:
Claudi, F., Tyson, A. L., Petrucco, L., Margrie, T.W., Portugues, R., Branco, T. (2021) "Visualizing anatomically registered data with Brainrender" <i>eLife</i> 2021;10:e65751 [doi.org/10.7554/eLife.65751](https://doi.org/10.7554/eLife.65751)
BibTeX:
@article{Claudi2021,
author = {Claudi, Federico and Tyson, Adam L. and Petrucco, Luigi and Margrie, Troy W. and Portugues, Ruben and Branco, Tiago},
doi = {10.7554/eLife.65751},
issn = {2050084X},
journal = {eLife},
pages = {1--16},
pmid = {33739286},
title = {{Visualizing anatomically registered data with brainrender}},
volume = {10},
year = {2021}
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file brainrender-2.1.17.tar.gz.
File metadata
- Download URL: brainrender-2.1.17.tar.gz
- Upload date:
- Size: 39.6 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.1.0 CPython/3.12.9
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
2e01212f4a891842555deff853c7e60be2628564d0ecf5fd864866166ec47aee
|
|
| MD5 |
bcc36f4299d3f4329be183c2a9b4b6fc
|
|
| BLAKE2b-256 |
581e43b70ddce036b70494a7010107cb9f2f236c4796c99012c8159953876f7b
|
File details
Details for the file brainrender-2.1.17-py3-none-any.whl.
File metadata
- Download URL: brainrender-2.1.17-py3-none-any.whl
- Upload date:
- Size: 43.3 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.1.0 CPython/3.12.9
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
d481b2268c2963fdb4dfb60fa6bc85a103c8e9204ab68c2ec6127854d577f088
|
|
| MD5 |
12c9d2db4a52691fc57510305e3b5709
|
|
| BLAKE2b-256 |
b133571a72490342f15501c73f128a207a2eb2b8eeb6461e7ea626e425260d28
|