A Python package for the identification, characterization and comparison of spatial clusters from spatial -omics data.
Project description
A Python package for the identification, characterization and comparison of spatial clusters from spatial omics data.
Documentation • Examples • Paper • Preprint
Background
Spatial clustering (or spatial domain identification) determines cellular niches characterized by specific admixing of these populations. It assigns cells to clusters based on both their intrinsic features (e.g., protein or mRNA expression), and the features of neighboring cells in the tissue.
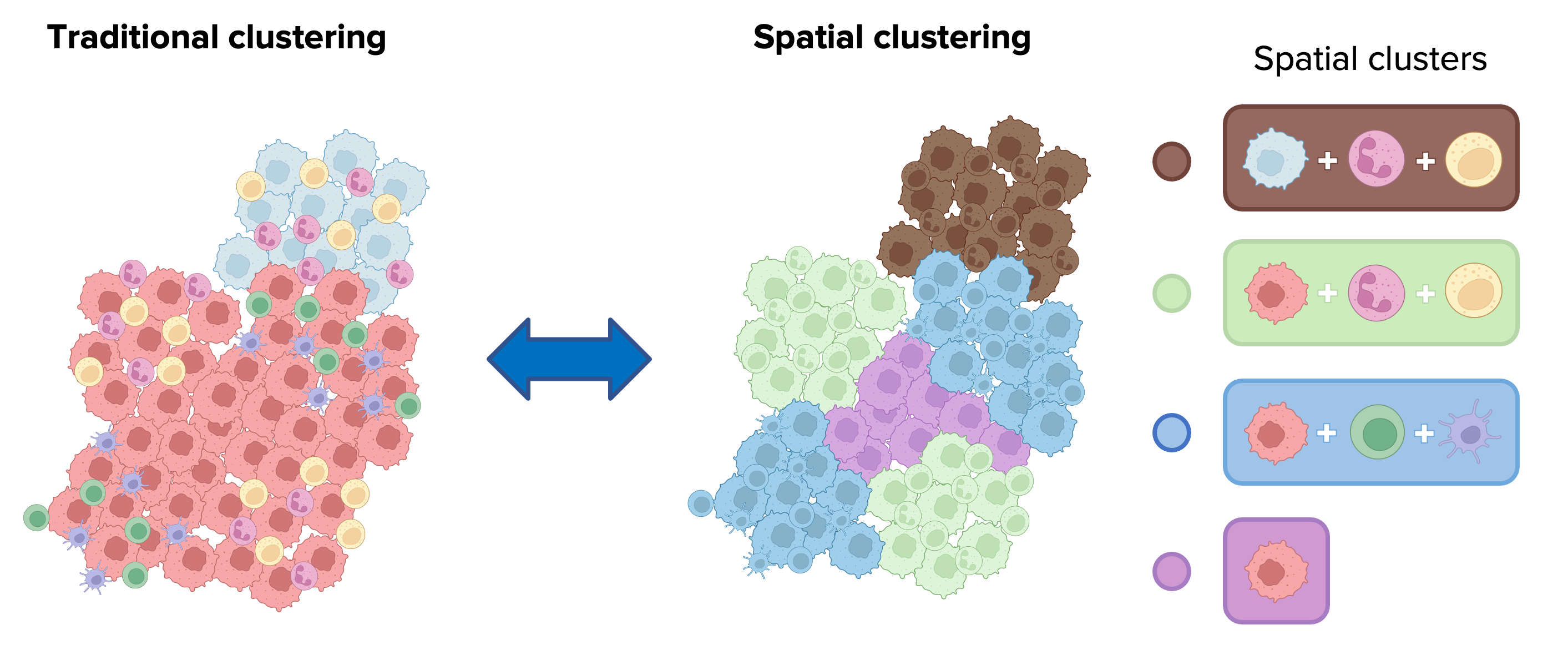
CellCharter is able to automatically identify spatial domains and offers a suite of approaches for cluster characterization and comparison.
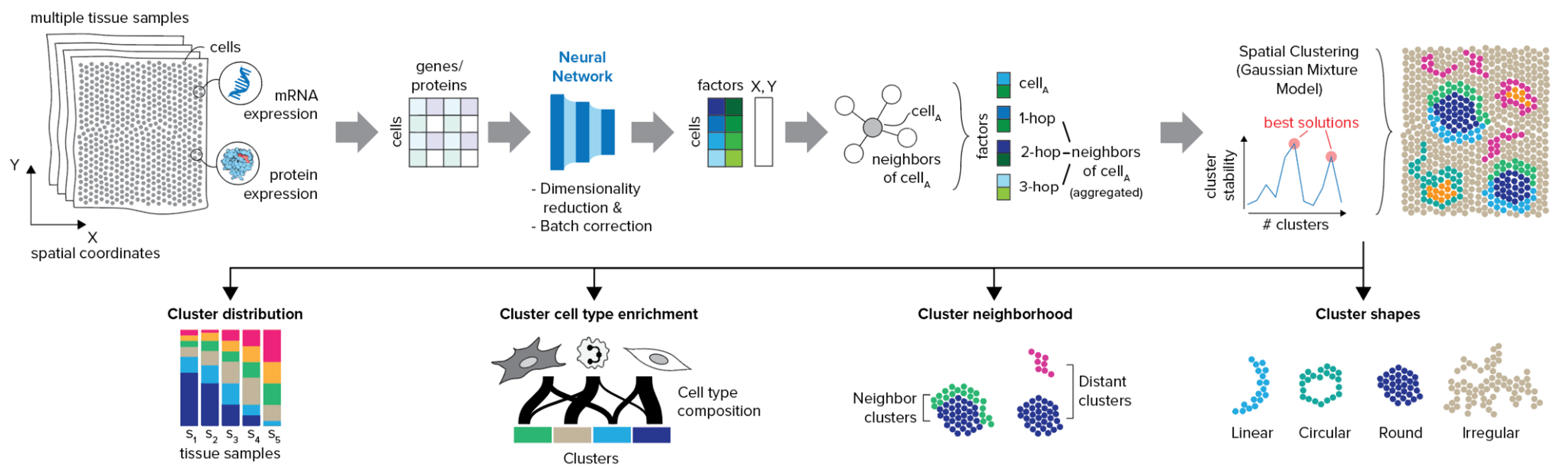
Features
- Identify niches for multiple samples: By combining the power of scVI and scArches, CellCharter can identify domains for multiple samples simultaneously, even with in presence of batch effects.
- Scalability: CellCharter can handle large datasets with millions of cells and thousands of features. The possibility to run it on GPUs makes it even faster
- Flexibility: CellCharter can be used with different types of spatial omics data, such as spatial transcriptomics, proteomics, epigenomics and multiomics data. The only difference is the method used for dimensionality reduction and batch effect removal.
- Spatial transcriptomics: CellCharter has been tested on scVI with Zero-inflated negative binomial distribution.
- Spatial proteomics: CellCharter has been tested on a version of scArches, modified to use Mean Squared Error loss instead of the default Negative Binomial loss.
- Spatial epigenomics: CellCharter has been tested on scVI with Poisson distribution.
- Spatial multiomics: it's possible to use multi-omics models such as MultiVI, or use the concatenation of the results from the different models.
- Best candidates for the number of domains: CellCharter offers a method to find multiple best candidates for the number of domains, based on the stability of a certain number of domains across multiple runs.
- Domain characterization: CellCharter provides a set of tools to characterize and compare the spatial domains, such as domain proportion, cell type enrichment, (differential) neighborhood enrichment, and domain shape characterization.
Since CellCharter 0.3.0, we moved the implementation of the Gaussian Mixture Model (GMM) from PyCave, not maintained anymore, to TorchGMM, a fork of PyCave maintained by the CSOgroup. This change allows us to have a more stable and maintained implementation of GMM that is compatible with the most recent versions of PyTorch.
Getting started
Please refer to the documentation. In particular, the
Installation
- Create a conda or pyenv environment
- Install Python >= 3.10,<3.13 and PyTorch >= 1.12.0. If you are planning to use a GPU, make sure to download and install the correct version of PyTorch first from here.
- Install the library used for dimensionality reduction and batch effect removal according to the data type you are planning to analyze:
- scVI for spatial transcriptomics and/or epigenomics data such as 10x Visium and Xenium, Nanostring CosMx, Vizgen MERSCOPE, Stereo-seq, DBiT-seq, MERFISH and seqFISH data.
- A modified version of scArches's TRVAE model for spatial proteomics data such as Akoya CODEX, Lunaphore COMET, CyCIF, IMC and MIBI-TOF data.
- Install CellCharter using pip:
pip install cellcharter
We report here an example of an installation aimed at analyzing spatial transcriptomics data (and thus installing scvi-tools).
This example is based on a Linux CentOS 7 system with an NVIDIA A100 GPU.
It will install Pytorch for GPU by default.
conda create -n cellcharter-env -c conda-forge python=3.12
conda activate cellcharter-env
pip install scvi-tools
pip install cellcharter
Note: a different system may require different commands to install PyTorch and JAX. Refer to their respective documentation for more details.
Contribution
If you found a bug or you want to propose a new feature, please use the issue tracker.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file cellcharter-0.3.5.tar.gz.
File metadata
- Download URL: cellcharter-0.3.5.tar.gz
- Upload date:
- Size: 44.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.12.9
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
3d792471b12750aae2d0e48aa855787b2327823dce9788646f4c0f7596d5387b
|
|
| MD5 |
24333ebc9373d7be93afb2a064222d53
|
|
| BLAKE2b-256 |
aa5166cd15011b448e280789250dda37be2a025424000067cb121a388ea77320
|
Provenance
The following attestation bundles were made for cellcharter-0.3.5.tar.gz:
Publisher:
release.yaml on CSOgroup/cellcharter
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
cellcharter-0.3.5.tar.gz -
Subject digest:
3d792471b12750aae2d0e48aa855787b2327823dce9788646f4c0f7596d5387b - Sigstore transparency entry: 253309082
- Sigstore integration time:
-
Permalink:
CSOgroup/cellcharter@5787208fec06fcfe92d8467e19e45eb200a817fe -
Branch / Tag:
refs/tags/v0.3.5 - Owner: https://github.com/CSOgroup
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
release.yaml@5787208fec06fcfe92d8467e19e45eb200a817fe -
Trigger Event:
release
-
Statement type:
File details
Details for the file cellcharter-0.3.5-py3-none-any.whl.
File metadata
- Download URL: cellcharter-0.3.5-py3-none-any.whl
- Upload date:
- Size: 49.4 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.12.9
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
c8fe08da4930704bc10132ca758e7a8c1725e24886f68aaf8f6b968745b2d697
|
|
| MD5 |
94db554b2269b301e1a6b4d6812497a2
|
|
| BLAKE2b-256 |
18178b846ec90e02903fdb530ff6b71e31b8c59d6d8d4bad58bc8947be9941f9
|
Provenance
The following attestation bundles were made for cellcharter-0.3.5-py3-none-any.whl:
Publisher:
release.yaml on CSOgroup/cellcharter
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
cellcharter-0.3.5-py3-none-any.whl -
Subject digest:
c8fe08da4930704bc10132ca758e7a8c1725e24886f68aaf8f6b968745b2d697 - Sigstore transparency entry: 253309084
- Sigstore integration time:
-
Permalink:
CSOgroup/cellcharter@5787208fec06fcfe92d8467e19e45eb200a817fe -
Branch / Tag:
refs/tags/v0.3.5 - Owner: https://github.com/CSOgroup
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
release.yaml@5787208fec06fcfe92d8467e19e45eb200a817fe -
Trigger Event:
release
-
Statement type:














