No project description provided
Project description
CNV From BAM
cnv_from_bam is a Rust library developed to efficiently calculate dynamic Copy Number Variation (CNV) profiles from sequence alignments contained in BAM files. It seamlessly integrates with Python using PyO3, making it an excellent choice for bioinformatics workflows involving genomic data analysis.
Features
- Efficient Processing: Optimized for handling large genomic datasets in BAM format.
- Python Integration: Built with PyO3 for easy integration into Python-based genomic analysis workflows.
- Multithreading Support: Utilizes Rust's powerful concurrency model for improved performance.
- Dynamic Binning: Bins the genome dynamically based on total read counts and genome length.
- CNV Calculation: Accurately calculates CNV values for each bin across different contigs.
- Directory Support: Supports processing of multiple BAM files in a directory. (Requires alignment to the same reference in all BAM files)
Installation
To use cnv_from_bam in your Rust project, add the following to your Cargo.toml file:
[dependencies]
cnv_from_bam = "0.1.0" # Replace with the latest version
Usage
Here's a quick example of how to use the iterate_bam_file function:
use cnv_from_bam::iterate_bam_file;
use std::path::PathBuf;
let bam_path = PathBuf::from("path/to/bam/file.bam");
// Iterate over the BAM file and calculate CNV values for each bin. Number of threads is set to 4 and mapping quality filter is set to 60.
// If number of threads is not specified, it defaults to the number of logical cores on the machine.
let result = iterate_bam_file(bam_path, Some(4), Some(60));
// Process the result...
The results in this case are returned as a CnvResult, which has the following structure:
pub struct CnvResult {
pub cnv: FnvHashMap<String, Vec<f64>>,
pub bin_width: usize,
pub genome_length: usize,
}
Where result.cnv is a hash map containing the Copy Number for each bin of bin_width bases for each contig in the reference genome, result.bin_width is the width of the bins in bases, and result.genome_length is the total length of the genome.
[!NOTE] Note: Only the main primary mapping alignment start is binned, Supplementary and Secondary alignments are ignored.
Directory analysis
To analyse a directory of BAM files, use the iterate_bam_dir function:
use cnv_from_bam::iterate_bam_dir;
use std::path::PathBuf;
let bam_path = PathBuf::from("path/to/bam_directory/");
// Iterate over the BAM files in teh directory and calculate CNV values for the whole. Number of threads is set to 4 and mapping quality filter is set to 60.
// If number of threads is not specified, it defaults to the number of logical cores on the machine.
let result = iterate_bam_file(bam_path, Some(4), Some(60));
This again returns a CnvResult, but this time the CNV values are summed across all BAM files in the directory. The bin width and genome length are calculated based on the first BAM file in the directory.
[!NOTE] Note: All BAM files in the directory must be aligned to the same reference genome.
Python Integration
cnv_from_bam can be used in Python using the PyO3 bindings. To install the Python bindings, run:
pip install cnv_from_bam
The same iterate_bam_file is available in python, accepting a path to a BAM file or a directory of BAM files, the number of threads (set to None to use the optimal number of threads for the machine), and the mapping quality filter.
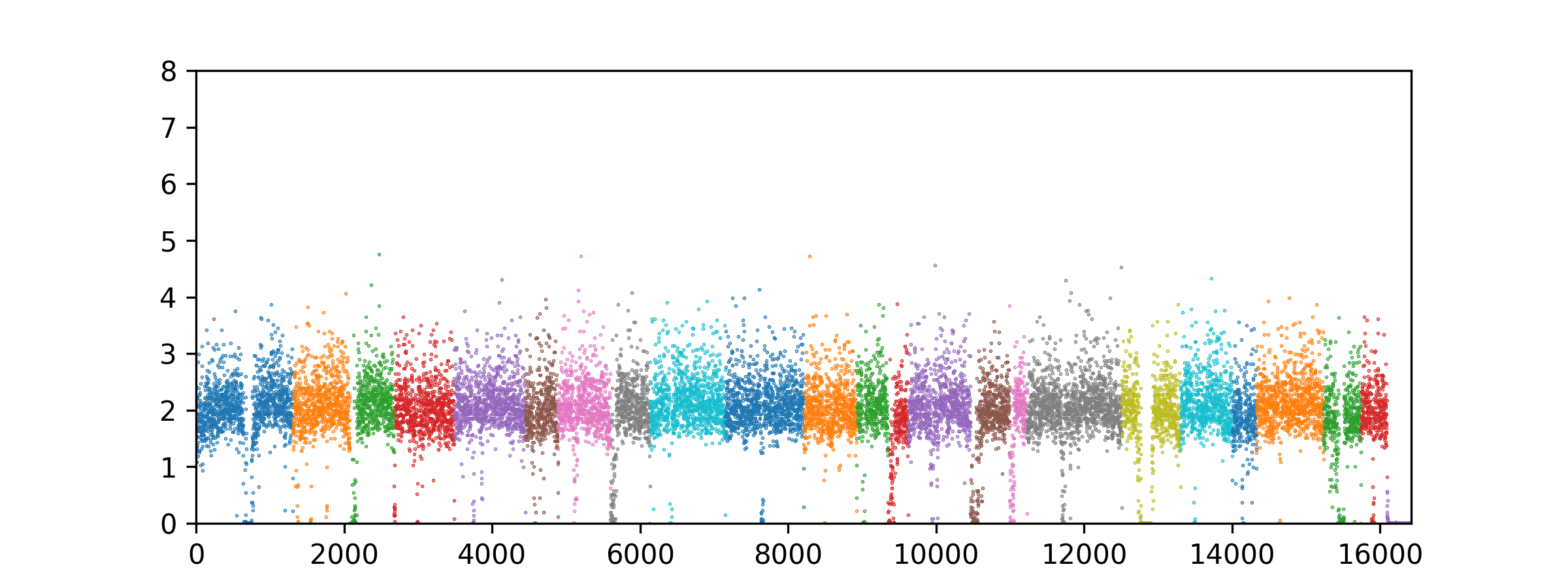
Example simple plot in python
```python
from matplotlib import pyplot as plt
import matplotlib as mpl
from pathlib import Path
import numpy as np
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(8, 3))
total = 0
bam_path = Path("path/to/bam/file.bam");
# Iterate over the BAM file and calculate CNV values for each bin. Number of threads is set to 4 and mapping quality filter is set to 60.
# If number of threads is not specified, it defaults to the optimal number of threads for the machine.
result = iterate_bam_file(bam_path, _threads=4, mapq_filter=60);
for contig, cnv in result.cnv.items():
ax.scatter(x=np.arange(len(cnv)) + total, y=cnv, s =0.1)
total += len(cnv)
ax.set_ylim((0,8))
ax.set_xlim((0, total))
Should look something like this. Obviously the cnv data is just a dictionary of lists, so you can do whatever you want with it vis a vis matplotlib, seaborn, etc.
Documentation
To generate the documentation, run:
cargo doc --open
Contributing
Contributions to cnv_from_bam are welcome!
We use pre-commit hooks (particularly cargo-fmt and ruff) to ensure that code is formatted correctly and passes all tests before being committed. To install the pre-commit hooks, run:
git clone https://github.com/Adoni5/cnv_from_bam.git
cd cnv_from_bam
pip install -e .[dev]
pre-commit install -t pre-commit -t post-checkout -t post-merge
pre-commit run --all-files
## License
This project is licensed under the [Mozilla Public License 2.0](LICENSE).
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distributions
Hashes for cnv_from_bam-0.2.0-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 4028b821c1d44940c0744384432d8b97deedde4134bd793a9d8e4906d20c742a |
|
| MD5 | 3da71e4b91c4b08d3dd99f2370f314b1 |
|
| BLAKE2b-256 | 56fe9b88b7aed24c4d5d387c934a27b9278fdd73adb8485638555ef627371877 |
Hashes for cnv_from_bam-0.2.0-pp310-pypy310_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 4b9b3b4a93ec494bda7d9373ea3c458b696fd0f499e53a54ff44d6713eaaf73a |
|
| MD5 | 97209159fe3f95284e0f0afc880c539b |
|
| BLAKE2b-256 | f0aac87f3955f531efd9405fd5d515183f1fe7fe0d86448323762b66b7a838ec |
Hashes for cnv_from_bam-0.2.0-pp310-pypy310_pp73-macosx_11_0_arm64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 5018b19566261550fca7120d4eb23265a8dcaba0c1687d7fe7c5f5316159b883 |
|
| MD5 | e9ed88ac619c923f189557ffa376ef1e |
|
| BLAKE2b-256 | 2d4ef3bf4e5b36a7f404802048174b78ac2310678f176c4656eeae7293b47b9a |
Hashes for cnv_from_bam-0.2.0-pp310-pypy310_pp73-macosx_10_7_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 979b20f38a052c364e2e37c0536562a7cea87d55f79428061da725a06b65f32d |
|
| MD5 | bb4ecec9a7b7191fb260fee9142a2168 |
|
| BLAKE2b-256 | 00847d3347b2795cd56a6b34324264bcd3bdfe098857706bb4e62740458f5908 |
Hashes for cnv_from_bam-0.2.0-pp39-pypy39_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | aeac39d3f72aa1af7aab33f726d4014be7644aa2aab2f4416012687cc31a8d07 |
|
| MD5 | 6c9ba43871bff6342813d92d35103702 |
|
| BLAKE2b-256 | 1b989e76f52da2173c203bfffa228ff89cc248b4bceed9b8985d847086f77283 |
Hashes for cnv_from_bam-0.2.0-pp39-pypy39_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 9f476181e0437ab85bd95ed61f5a41a1c269d77b3b02862088815bcc5f0695a1 |
|
| MD5 | c72573fa3fdbe031b97ef6e3f2dc5d79 |
|
| BLAKE2b-256 | fb86974c0ead3c29b3c2a35c6e7f16d917b257fc67bbe1703cb252d5fec72748 |
Hashes for cnv_from_bam-0.2.0-pp39-pypy39_pp73-macosx_11_0_arm64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 6be296f1ce4f6d43a6f3d7f6dc4f9c2866a7129376ac59a1c0b7856ba738bb0d |
|
| MD5 | 3f0d37506493baab48c3c55fa84862f8 |
|
| BLAKE2b-256 | d1efa2eaa47f97139585b42a807b29ff320748d328c916965c1a3792daa7efad |
Hashes for cnv_from_bam-0.2.0-pp39-pypy39_pp73-macosx_10_7_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | dcefb57c1efd130ae46f4e6292f664b6a64993c6e63d47a1a49e04a5c2e7092e |
|
| MD5 | d3a1eed6a3625b2091066c6cd622d01e |
|
| BLAKE2b-256 | 198eb1669a4e508881e2f089310f6454578bfa475abfebde62dbce5b49c0cee8 |
Hashes for cnv_from_bam-0.2.0-pp38-pypy38_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 8afdc0c88be0c8de92fe3ae495da90c0bd67732be44beeb3197bf3281ef3b9ab |
|
| MD5 | 5029b8a34a0cd6374c94560f53cf5443 |
|
| BLAKE2b-256 | df87464602dda5d970105bd8b4102b1b1c918c4977cd89c2f58dab0b61cd5563 |
Hashes for cnv_from_bam-0.2.0-pp38-pypy38_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | fc00ceb0e6005872ba81a5526b14ba547fad0923849ef72f8843afe85efc4d1d |
|
| MD5 | de1b7766693327cf1ac0acf44c55994c |
|
| BLAKE2b-256 | 573ec7cf96b3602b67f40962fcb07f98676ece9e1df1075e80285ef9d8e506d2 |
Hashes for cnv_from_bam-0.2.0-pp38-pypy38_pp73-macosx_11_0_arm64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 1503d0049b391db44f9900c460281ca5889f40397747c218e4a60e3852fa987f |
|
| MD5 | d6b2133ffc73f3d6e60a450663d40068 |
|
| BLAKE2b-256 | f8aba992992002f6afe980ae17fb57096094affb5cd343a9868bfc44e3f31054 |
Hashes for cnv_from_bam-0.2.0-pp38-pypy38_pp73-macosx_10_7_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 8c1217817ffce03bbe8f743d1f3334c579b8c79db9a167dde2b2e796e3785fb0 |
|
| MD5 | 6b9ad173a60cb50258a441148db4f690 |
|
| BLAKE2b-256 | 33ad914ae78f529b8bb9fb3aa2fcf748bab6ee167541888f0e3db1c64834145f |
Hashes for cnv_from_bam-0.2.0-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 347ffe4b6691d544a00fcdd8058ef8a29ee7214ac1e6048b7a24d2e993fd212a |
|
| MD5 | 1e23528fbd74025ad78f219f541b871a |
|
| BLAKE2b-256 | 3a9ab94e73afe49d432d29e052a35113e84bc60737b81c5051c7f6046a66f308 |
Hashes for cnv_from_bam-0.2.0-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 707b9f54ec767b85fb0707a2f83ebc71d3e08b290bb00711d27d0e7b3d9bf3bd |
|
| MD5 | 10378987d3fb6b221588b264b20ea16c |
|
| BLAKE2b-256 | 693260cb17f55ef5fd27e264760b8440957f1c3dc2f2a12d02339aad2ec2feb7 |
Hashes for cnv_from_bam-0.2.0-cp312-cp312-macosx_11_0_arm64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | ea020ed227e7fbe85ae68d4cfcfa95c50df2b3267e44a8551882fe4ec32c4adb |
|
| MD5 | 50026b6da0b9a0b531f652d20f3f0b0e |
|
| BLAKE2b-256 | 91aa4c383c10e51b99012f321b0ba97caef609f62696a95a7c34a6a876170ca5 |
Hashes for cnv_from_bam-0.2.0-cp312-cp312-macosx_10_7_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 2800ecfa51548af61dd347fc2ac83e1338b27cf2e3f1d3ef573b507794cf44f4 |
|
| MD5 | f4b038d92538af65e0e5f59675919be7 |
|
| BLAKE2b-256 | 0a9d2c02cc275a7f1c2eb1e66e4fbefe5bed0ab8c7a94fbf84c28fa3240a6a9b |
Hashes for cnv_from_bam-0.2.0-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 991220eeb82a17227036b080e357e6c55e28ad9c891c9117aadb5a44c7faeb87 |
|
| MD5 | 62deee3193e2ae0c82a193d4bb2d7a1f |
|
| BLAKE2b-256 | b929bdb12fe04fcd78a730825c9a73986fa900dbf24b98820f69e9c977a9829b |
Hashes for cnv_from_bam-0.2.0-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 017db38ff1bdb4e582d1acb1b2168a1b466203f6c531e83cd817064fb00ae360 |
|
| MD5 | 6a0f847af66d24821a953547743027b2 |
|
| BLAKE2b-256 | cb9bf6c4455241194c4852063afa997bf1ce47d0afa71ae84fad784da162a4ae |
Hashes for cnv_from_bam-0.2.0-cp311-cp311-macosx_11_0_arm64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 357936d8f260ff2c53319d3551824948a12c09bc8ad635c48d0bed8efe5f634c |
|
| MD5 | cb0e5862bef6a7423f1a227b025fe2e2 |
|
| BLAKE2b-256 | 54ea60a312695b863b8a8ab2c02abff7d7e144094e6aaa2afccedd59cd50bb8d |
Hashes for cnv_from_bam-0.2.0-cp311-cp311-macosx_10_7_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | d903343a6b58060b691b596de3462246681edb82f7743f9f27f3bafff47866e8 |
|
| MD5 | d9d26a54a211dc0971b64cae54cd9c32 |
|
| BLAKE2b-256 | bdc8c99ceb7213c11dfc34810cffc70dfaf80f2b6b4188825400a1cea3e20095 |
Hashes for cnv_from_bam-0.2.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | ffdfe0b3609a20833d259d04d25ebf6799eed92a65653f2181ae8acb6c091a2d |
|
| MD5 | 6cd72394be33f0dad8ce19eb706f2afc |
|
| BLAKE2b-256 | f91772a66d9865154df99609e0b17bf490111a67222b7b882b098c29d6e60253 |
Hashes for cnv_from_bam-0.2.0-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 7c7ee4796facee8462713d0b45cdb4f30660c8c2a5f54ea921bd24d2f7a03bd1 |
|
| MD5 | 471fde8b5adc1c1b4cab1d9b5e7cd13e |
|
| BLAKE2b-256 | c22a264e06aebc3b9aaad0f204e99b6dc24c90608d68f8377b8b0e9c8d029446 |
Hashes for cnv_from_bam-0.2.0-cp310-cp310-macosx_11_0_arm64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | f9f4eee443edba30a376e0eecb43f00fa0d8705d6fa917a5d025d51ee7f57716 |
|
| MD5 | bae8ec2383978e482e19aa3b212918dc |
|
| BLAKE2b-256 | 76224032f3dc2da52c97f4a0d333803413ef4f0514499b2e590c2e0e2251d7a9 |
Hashes for cnv_from_bam-0.2.0-cp310-cp310-macosx_10_7_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | bd8b724395439abe67a2a89f6700f9be8a4a92449ff6b8df5fdaba19fa98b49d |
|
| MD5 | 0842b5bcc86dd110546daa2fbd053c91 |
|
| BLAKE2b-256 | 41e101013da6571e5e4b2e6eb9b31297144c3062c31a9dcb466c3fae8b7e051c |
Hashes for cnv_from_bam-0.2.0-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | afc5b8b663db856ce28340e76ca5ac961959fc748f82aebbb18759298351638f |
|
| MD5 | 8850596eb731d214acde9a12b862ed76 |
|
| BLAKE2b-256 | 11c3121c271aefc0d9a536013628bc8cdc23b494141f44d3dac790d07fdd58b0 |
Hashes for cnv_from_bam-0.2.0-cp39-cp39-manylinux_2_17_aarch64.manylinux2014_aarch64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 27058a94cd34fe2f940f28a7c4f47f3fef72f54ccff3a53c75da186bbda2c018 |
|
| MD5 | d317897fba0ddb42f82c824bde978b7a |
|
| BLAKE2b-256 | 21da1c2e8dfd17a370cb77d427759912394fd3c105259162c74c1b9b10e56a64 |
Hashes for cnv_from_bam-0.2.0-cp39-cp39-macosx_11_0_arm64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | abca4f5c0fcb4017a269bd54088c9ffaa93f925a4002cdc30196f5b58b26bb2b |
|
| MD5 | ddea95f52c84188040b2ad633733e920 |
|
| BLAKE2b-256 | c6a3f4c784bf6c400f016ec9d843be02137a49e1c5211f45abb01e17df34d8c7 |
Hashes for cnv_from_bam-0.2.0-cp39-cp39-macosx_10_7_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | e6880bba1e657dd4c19c0b25500623a760282ffa8b168a88e7c3e973b2d9692a |
|
| MD5 | 922c5bd3dcbee160f19968132efc796e |
|
| BLAKE2b-256 | 8e48fea1eb1bc66633b76f01f9fc4c72a0bb1d53a26bdb1c462ae4be779ad014 |
Hashes for cnv_from_bam-0.2.0-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 7954605f392f14ac1ee9a3ecac54834d7131162a111666b493087978a5aa5292 |
|
| MD5 | f7c3d923672f1f5bded8182f45c00629 |
|
| BLAKE2b-256 | a4dc7efc7d5d606f3beee9e2c47b9a43071175ab8927171500879ba7f3264879 |
Hashes for cnv_from_bam-0.2.0-cp38-cp38-manylinux_2_17_aarch64.manylinux2014_aarch64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | dba279e76cf4a1901b7cddb00553ed448b76696dfda819eeaed18025773b6c47 |
|
| MD5 | b7051c7cdc67a977f4db77f9ed696c59 |
|
| BLAKE2b-256 | 32ab0aa88cd46457ff04c9fca9191d12311aeae17250bf0784c8c7c6b996377d |
Hashes for cnv_from_bam-0.2.0-cp38-cp38-macosx_11_0_arm64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 1d765a6d61ebb85aa788facc79d87f1e8b9e62ad07050bb697c7fd6011af15a4 |
|
| MD5 | 8a9793cff4a833889562acb4b803b551 |
|
| BLAKE2b-256 | 15778134218e438649ff1056794d196aca226259c68f11b6ef5824f980659611 |
Hashes for cnv_from_bam-0.2.0-cp38-cp38-macosx_10_7_x86_64.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 6f5338e4bff239fb307054bebd5d23aee2fe03aac1bfe31796f40e2b9012c5b7 |
|
| MD5 | 44a4fdd6d181f4831726be7e3ecb9759 |
|
| BLAKE2b-256 | 741cdbdaba3eba7a62b5f8b7c0e38a991c8934923295e857e1996f445b206d98 |