Data loader part of the mid-project for the Data Science bootcamp from Core Code School
Project description
COVID Data Loader
This project is part of the Core Data COVID project. Here are the tools needed for:
- Create the database
- Populate the database
- Query the data
Table of contents
Installation 📥
Library 📖
To use this project as a library, you have to install it with pip:
pip install covid-data
CLI 🤖
To use it as CLI, you still have to install it with pip. After that, you can run the commands from a terminal:
covid-data --help
Configuration ⚙
The project looks for the following environment variables to configure several parts:
- POSTGRES_USER: Postgres username
- POSTGRES_PASS: Postgres password
- POSTGRES_HOST: Postgres host
- POSTGRES_PORT: Postgres port
- POSTGRES_DB: Postgres database
- CAGEDATA_API_KEY: OpenCageData API Key
You can create the OpenCage data ley from their website. The package uses it to enrich and normalize places.
Usage as CLI 🎛️
To populate the database, you may use the commands the library provides.
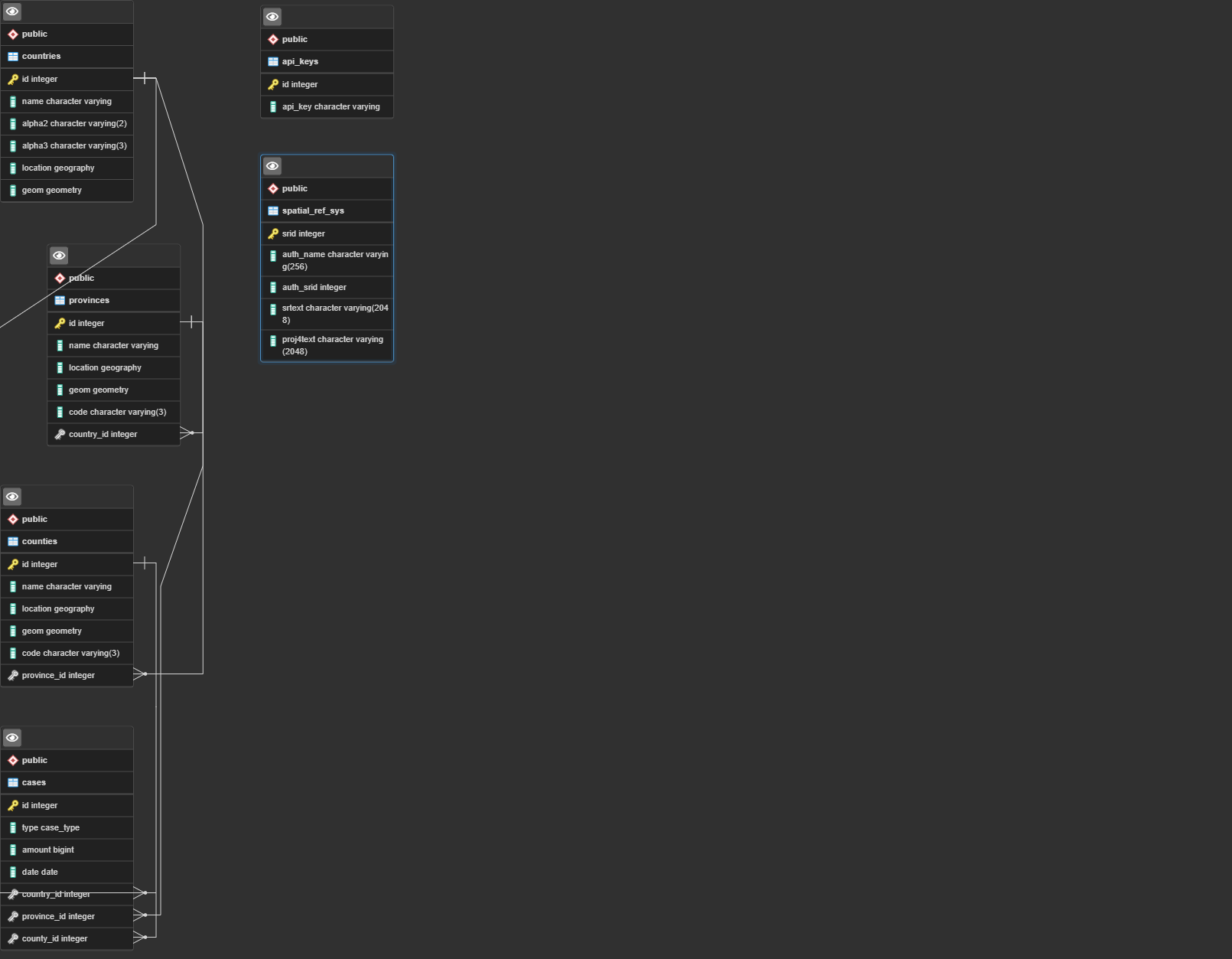
The first step is to create the database with the schema available here. The database structure is the following one:
Once the package finishes creating the database, you can populate it from a CSV with the command:
❯ covid-data loadcsv /path/to/confirmed.csv,/path/to/dead.csv,/path/to/recovered.csv -tf -o
For getting help on how to use the command, run:
❯ covid-data --help
Usage: covid-data [OPTIONS] COMMAND [ARGS]...
Options:
--help Show this message and exit.
Commands:
loads Loads FILES as CSV data.
scrap Scrap cases of chosen COUNTRY.
loadcsv
❯ covid-data loadcsv --help
Usage: covid-data loadcsv [OPTIONS] FILES
Loads FILES as CSV data. If you want to load several files, each file should be separated by a comma
Options:
-t, --type TEXT Type of cases contained in each file, separated by
comma. Leave blank if using --type-in-file
-tf, --type-in-file Set this to true if the file names are <case_type>.csv
Being <case_type> one of confirmed, recovered or dead
-o, --optimize Set to true to skip lines for places that has more
cases than columns on the CSV
--help Show this message and exit.
This command allows you to populate the database with one or more CSV files. Each file may only contain one case type.
You have to provide the paths separated by commas.
As well as with the file names, each file case type has to be separated by commas. Pass the type list under the argument --type and in the same order as the files.
Another option is not using the --type argument. In that case, you have to name the files according to the case type each of them contains (e.g., recovered.csv). If you choose this alternative, you have to set the flag --type-in-file.
The last flag is --optimize. If set, for each row, the CLI is going to get the country from that row. Then check the number of cases already saved for that country (with the same type as the file). If the country has the same or more number of cases as columns are in the CSV, it will skip that row.
An example of how to run the command for loading all CSVs:
❯ covid-data loadcsv /path/to/confirmed.csv,/path/to/dead.csv,/path/to/recovered.csv -tf -o
In that example, I'm using the file name to specify the case type contained. I'm also setting the optimization flag.
scrap
With this command, you can scrap the cases of a specific country, allowing you to extend the data from that country.
The command usage is as follows:
❯ covid-data scrap --help
Usage: covid-data scrap [OPTIONS] COUNTRY
Scrap cases of chosen COUNTRY. To check available countries to scrap use
--check
Options:
--check to Use this to check available countries
--start-date TEXT Date to start scraping cases from, in format DD/MM/YYYY
--help Show this message and exit.
There is one required argument, the country to scrap. If you want to know which countries are available, you can pass whatever value to COUNTRY and set the flag --check:
❯ covid-data scrap XXX --check
Available countries are:
France
Spain
Once you know which country to scrap, you can launch the command:
❯ covid-data scrap Spain
Fetching cases for province 1/20
Fetching cases for province 2/20
Fetching cases for province 3/20
...
You can set the start date with the argument --start-date. The date format required is DD/MM/YYYY
❯ covid-data scrap France --start-date 01/08/2021
Populate DB from backup
You may find attached to each release, a covid-data.sql file. That script allows you to load into an SQL database the most recent data.
If you run that script on the covid-data database, it will populate it with the necessary data for running the app.
Contributing ♥
You can become part of this project by proposing (and even implementing) enhancements, new commands, or new scrappers.
Add new scrappers
Adding a new scrapper is as simple as creating a new file named <country_name>.py in the folder covid_data/scrappers and make that file expose a function with the following signature:
def scrap(db_engine: psycopg2._psycopg.connection, start_date: datetime.datetime)
That function has to be in charge of scrapping, processing, formatting as well as saving new cases for the requested country.
Add new commands
Adding a new command is also a simple process. Place a file named <command_name>.py in the folder covid_data/commands and expose a function main with the needed click decorators.
A mandatory decorator is @click.command("command-name"), to avoid overwriting commands as all functions are named main.
As library
The potential of this library is offered by the db submodule, which you can find on CODE.
License
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file covid_data-0.1.21.tar.gz.
File metadata
- Download URL: covid_data-0.1.21.tar.gz
- Upload date:
- Size: 22.3 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.2 importlib_metadata/4.6.4 pkginfo/1.7.1 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.1 CPython/3.9.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
65195dc13d6ab577318a4586b828b0e6db95f75adef75cb9f11cc3dccd3fa988
|
|
| MD5 |
3685ece6439d135b6a600176b19399a6
|
|
| BLAKE2b-256 |
7e1d1b7ee2a2ca7a27906c97e65284bde83c760d96ca59c9f41e9c0b7dd19867
|
File details
Details for the file covid_data-0.1.21-py3-none-any.whl.
File metadata
- Download URL: covid_data-0.1.21-py3-none-any.whl
- Upload date:
- Size: 24.1 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.2 importlib_metadata/4.6.4 pkginfo/1.7.1 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.1 CPython/3.9.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
06c8f813f3d99990af5b9d7228ca073d8fe38ab663e46784443689f176674f7c
|
|
| MD5 |
5852c7ce94806043026849157b884ff0
|
|
| BLAKE2b-256 |
8c00c4d8e853dbc9829037353702494a5ca893e92b1db698aa569278359ca36e
|