A small package that implements the SEIRD Epidemiological Model on COVID-19 data.
Project description
covid-seird
About
covid-seird is a small Python package inspired by Henri Froese's post 'Infectious Disease Modelling: Beyond the Basic SIR Model'.
It implements the SEIRD Epidemiological Model on COVID-19 data.
- First, it fits a the SEIRD Model into the real timeline data of confirmed cases of COVID-19 of a country.
- Then, it simulates the SEIRD curves based on the previously fitted Model.
- As a result, the country's basic reproduction number (Ro) of COVID-19 is computed.
- Also, the fit and simulation plots are available.
NOTE: The COVID-19 timelines data are obtained using the COVID19Py package in order to retrieve the Worldwide Data repository operated by the Johns Hopkins University Center for Systems Science and Engineering (JHU CSSE).
Installation
In order to install this package, simply run:
pip install covid-seird
Usage
To use covid-seird, you first need to import the package and then create a new instance, passing the country code as a parameter:
from covid_seird import CountryCovidSeird
brazil = CountryCovidSeird("br")
Getting the country codes
CountryCovidSeird.code_search("brazil")
output:
{'BR': 'Brazil'}
Fit the SEIRD Model
brazil.fit()
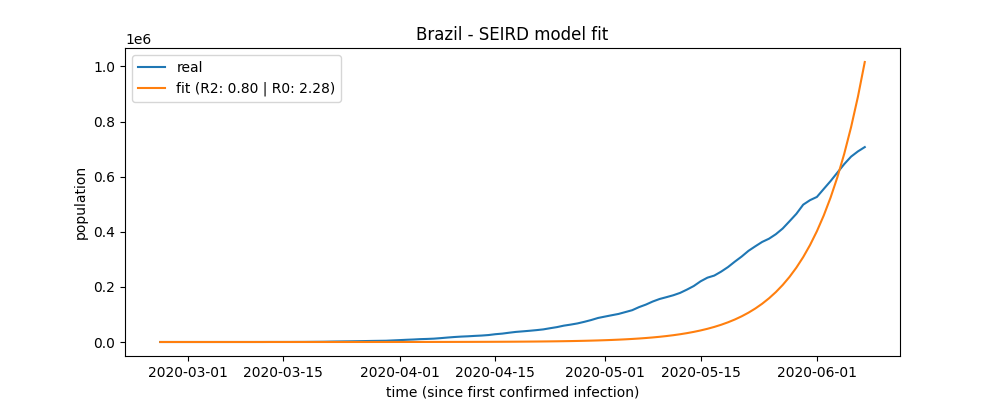
After the fit method's call, the fit score (R2) can be accessed:
brazil.r2
output:
0.9183167162166936
Also after fit method's call, the basic reproduction number (Ro) will be available:
brazil.r0
output:
2.2353912163446745
Fit plot
brazil.plot_fit("brazil_fit_plot")
a plot file named 'brazil_fit_plot' will be created.
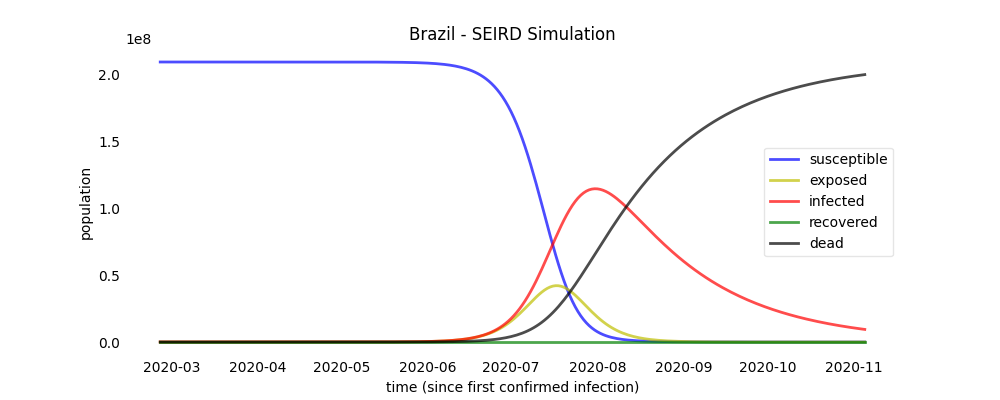
SEIRD simulation
The simulation method receives as a parameter how many days ahead of the real data the SEIRD model will be computed.
brazil.simulation(days_ahead=150)
The SEIRD curves can be accessed after the simulation method's call
brazil.curves['infected']
output:
array([4.77396851e-09, 5.32992037e-09, 5.95375686e-09, 6.59631344e-09,
7.32558927e-09, 8.14396099e-09, 9.05142859e-09, 1.00479921e-08,
...
7.48473200e-03, 7.06776956e-03, 6.67386382e-03, 6.30175876e-03,
5.95026480e-03, 5.61825534e-03, 5.30466353e-03, 5.00847963e-03])
Simulation plots
brazil.plot_simulation("brazil_simulation_plot")
a plot file named 'brazil_simulation_plot' will be created.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file covid-seird-0.0.6.tar.gz.
File metadata
- Download URL: covid-seird-0.0.6.tar.gz
- Upload date:
- Size: 5.3 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.1.1 pkginfo/1.5.0.1 requests/2.23.0 setuptools/47.1.1 requests-toolbelt/0.9.1 tqdm/4.46.0 CPython/3.6.9
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
55de4e7726864acb1af54124a4490e9a83b01bb81ee750b8e7b6d1c848eba9df
|
|
| MD5 |
78ac5e4428a64a019d10fc6f1a4729ec
|
|
| BLAKE2b-256 |
57945a34dbcc46617d2c45e07680de4f9601c9947588c20fee9a92ff299efba3
|
File details
Details for the file covid_seird-0.0.6-py3-none-any.whl.
File metadata
- Download URL: covid_seird-0.0.6-py3-none-any.whl
- Upload date:
- Size: 6.3 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.1.1 pkginfo/1.5.0.1 requests/2.23.0 setuptools/47.1.1 requests-toolbelt/0.9.1 tqdm/4.46.0 CPython/3.6.9
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
ac7d9a11717cd8705e8b571c54de5e96a9c1656b50ed6cce569a76b0d77296ea
|
|
| MD5 |
ae707b65258a05a7be23f40fdc527235
|
|
| BLAKE2b-256 |
4bf93e2bfa87fb3815ec71d735ae22ca71bb78af5821fcd36257e938929ec451
|