Implement DeepCAD-RT to denoise data by removing independent noise
Project description
DeepCAD-RT: Real-time denoising of fluorescence time-lapse imaging using deep self-supervised learning

Project page | Paper
Overview
Among the challenges of fluorescence microscopy, poor imaging signal-to-noise ratio (SNR) caused by limited photon budget lingeringly stands in the central position. Fluorescence microscopy is inherently sensitive to detection noise because the photon flux in fluorescence imaging is far lower than that in photography. For almost all fluorescence imaging technologies, the inherent shot-noise limit determines the upper bound of imaging SNR and restricts the imaging resolution, speed, and sensitivity. To capture enough fluorescence photons for satisfactory SNR, researchers have to sacrifice imaging resolution, speed, and even sample health.
We present a versatile method DeepCAD-RT to denoise fluorescence time-lapse images with rapid processing speed that can be incorporated with the microscope acquisition system to achieve real-time denoising. Our method is based on deep self-supervised learning and the original low-SNR data can be directly used for training convolutional networks, making it particularly advantageous in functional imaging where the sample is undergoing fast dynamics and capturing ground-truth data is hard or impossible. We have demonstrated extensive experiments including calcium imaging in mice, zebrafish, and flies, cell migration observations, and the imaging of a new genetically encoded ATP sensor, covering both 2D single-plane imaging and 3D volumetric imaging. Qualitative and quantitative evaluations show that our method can substantially enhance fluorescence time-lapse imaging data and permit high-sensitivity imaging of biological dynamics beyond the shot-noise limit.
For more details, please see the companion paper where the method first appeared: "Real-time denoising of fluorescence time-lapse imaging enables high-sensitivity observations of biological dynamics beyond the shot-noise limit, bioRxiv (2022)".
Pytorch code
Our environment
- Ubuntu 16.04
- Python 3.6
- Pytorch 1.8.0
- NVIDIA GPU (GeForce RTX 3090) + CUDA (11.1)
Environment configuration
-
Create a virtual environment and install PyTorch. In the 3rd step, please select the correct Pytorch version that matches your CUDA version from https://pytorch.org/get-started/previous-versions/.
$ conda create -n deepcadrt python=3.6 $ conda activate deepcadrt $ pip install torch==1.8.0+cu111 torchvision==0.9.0+cu111 torchaudio==0.8.0 -f https://download.pytorch.org/whl/torch_stable.htmlNote:
pip installcommand is required for Pytorch installation. -
We made a installable pip realease of DeepCAD-RT [pypi]. You can install it by entering the following command:
$ pip install deepcad
Download the source code
$ git clone https://github.com/cabooster/DeepCAD-RT
$ cd DeepCAD-RT/DeepCAD_RT_pytorch/
Demos
To try out the Python code, please activate the deepcadrt environment first:
$ conda activate deepcadrt
$ cd DeepCAD-RT/DeepCAD_RT_pytorch/
Example training
To train a DeepCAD-RT model, we recommend starting with the demo script demo_train_pipeline.py. One demo dataset will be downloaded to the DeepCAD_RT_pytorch/datasets folder automatically. You can also download other data from the companion webpage or use your own data by changing the training parameter datasets_path.
python demo_train_pipeline.py
Example testing
To test the denoising performance with pre-trained models, you can run the demo script demo_test_pipeline.py . A demo dataset and its denoising model will be automatically downloaded to DeepCAD_RT_pytorch/datasets and DeepCAD_RT_pytorch/pth, respectively. You can change the dataset and the model by changing the parameters datasets_path and denoise_model.
python demo_test_pipeline.py
Jupyter notebook
We provide simple and user-friendly Jupyter notebooks to implement DeepCAD-RT. They are in the DeepCAD_RT_pytorch/notebooks folder. Before you launch the notebooks, please configure an environment following the instruction in Environment configuration . And then, you can launch the notebooks through the following commands:
$ conda activate deepcadrt
$ cd DeepCAD-RT/DeepCAD_RT_pytorch/notebooks
$ jupyter notebook
Colab notebook
We also provide a cloud-based notebook implemented with Google Colab. You can run DeepCAD-RT directly in your browser using a cloud GPU without configuring the environment.
Note: The Colab notebook needs much longer time to train and test because of the limited GPU performance offered by Colab.
Matlab GUI
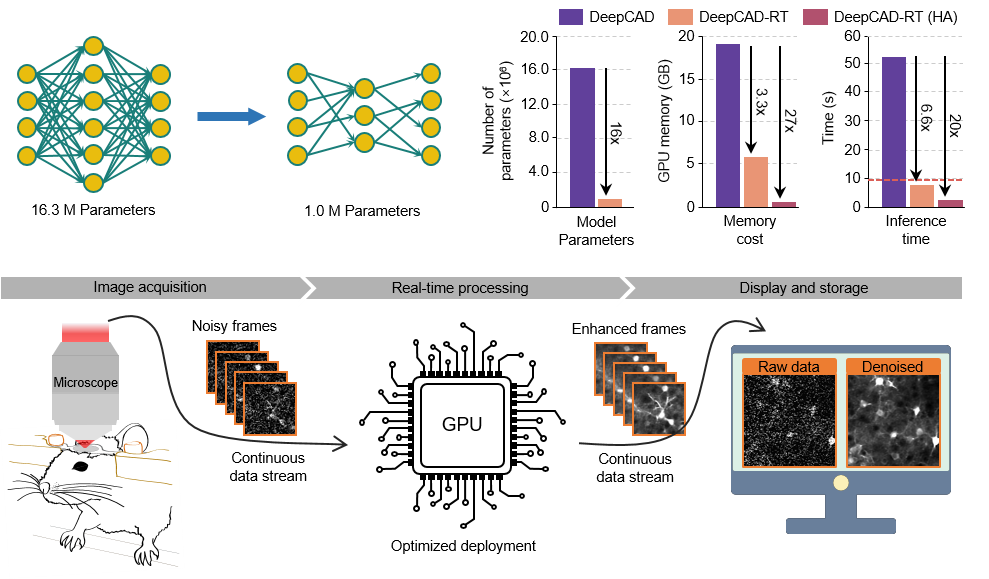
To achieve real-time denoising, DeepCAD-RT was optimally deployed on GPU using TensorRT (Nvidia) for further acceleration and memory reduction. We also designed a sophisticated time schedule for multi-thread processing. Based on a two-photon microscope, real-time denoising has been achieved with our Matlab GUI of DeepCAD-RT (tested on a Windows desktop with Intel i9 CPU and 128 GB RAM). Tutorials on installing and using the GUI has been moved to this page.

Results
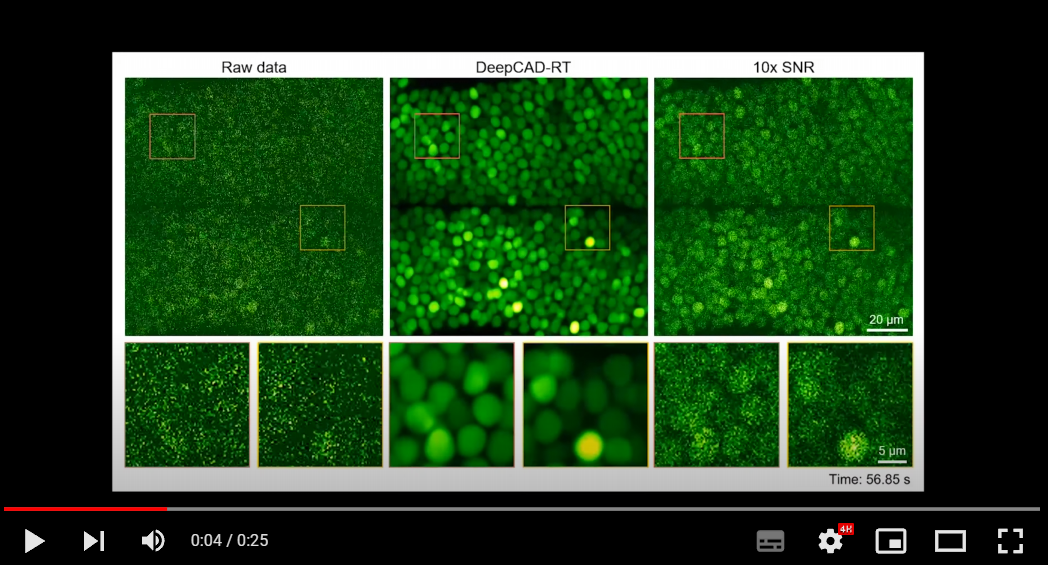
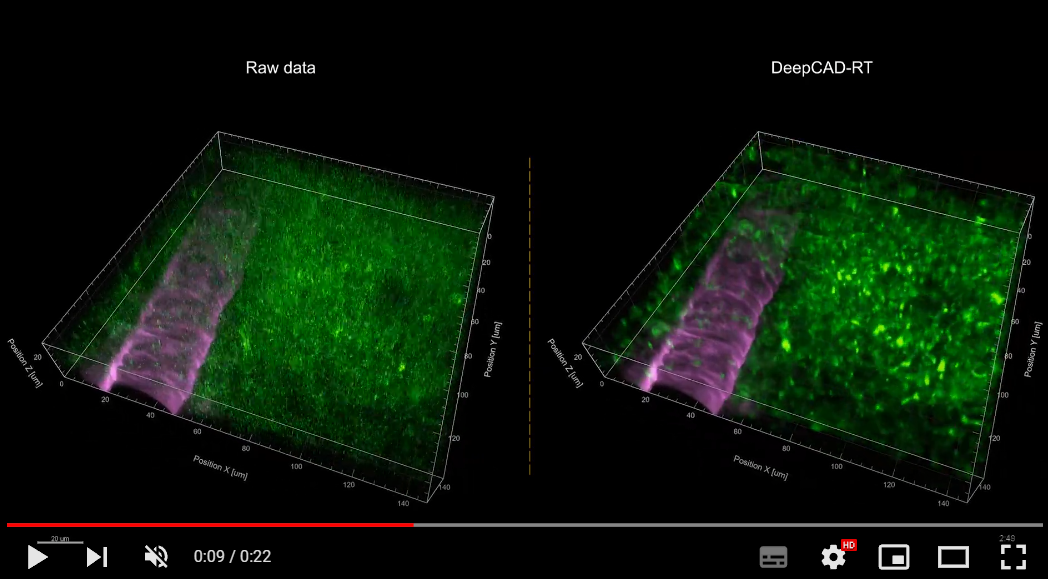
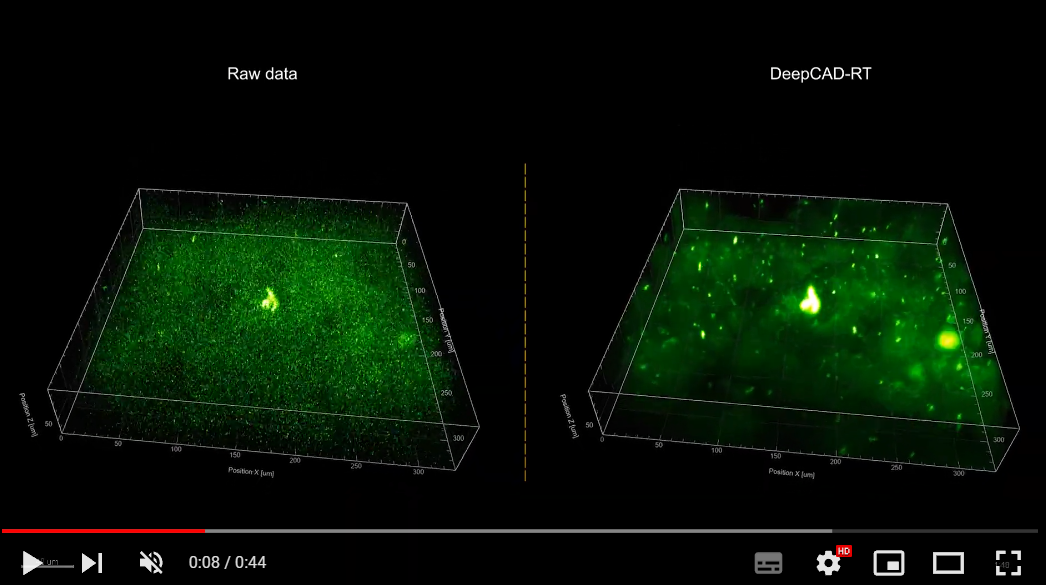
1. Universal denoising for calcium imaging in zebrafish.
2. Denoising performance of DeepCAD-RT of neutrophils in the mouse brain in vivo.
3. Denoising performance of DeepCAD-RT on a recently developed genetically encoded ATP sensor.
More demo videos are demonstrated on our website.
Citation
If you use this code please cite the companion paper where the original method appeared:
-
Xinyang Li, Yixin Li, Yiliang Zhou, et al. Real-time denoising of fluorescence time-lapse imaging enables high-sensitivity observations of biological dynamics beyond the shot-noise limit. bioRxiv (2022). https://doi.org/10.1101/2022.03.14.484230
-
Xinyang Li, Guoxun Zhang, Jiamin Wu, et al. Reinforcing neuron extraction and spike inference in calcium imaging using deep self-supervised denoising. Nat Methods 18, 1395–1400 (2021). https://doi.org/10.1038/s41592-021-01225-0
@article {Li2022.03.14.484230,
author = {Li, Xinyang and Li, Yixin and Zhou, Yiliang and Wu, Jiamin and Zhao, Zhifeng and Fan, Jiaqi and Deng, Fei and Wu, Zhaofa and Xiao, Guihua and He, Jing and Zhang, Yuanlong and Zhang, Guoxun and Hu, Xiaowan and Zhang, Yi and Qiao, Hui and Xie, Hao and Li, Yulong and Wang, Haoqian and Fang, Lu and Dai, Qionghai},
title = {Real-time denoising of fluorescence time-lapse imaging enables high-sensitivity observations of biological dynamics beyond the shot-noise limit},
elocation-id = {2022.03.14.484230},
year = {2022},
doi = {10.1101/2022.03.14.484230},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2022/03/14/2022.03.14.484230},
eprint = {https://www.biorxiv.org/content/early/2022/03/14/2022.03.14.484230.full.pdf},
journal = {bioRxiv}
}
@article{li2021reinforcing,
title={Reinforcing neuron extraction and spike inference in calcium imaging using deep self-supervised denoising},
author={Li, Xinyang and Zhang, Guoxun and Wu, Jiamin and Zhang, Yuanlong and Zhao, Zhifeng and Lin, Xing and Qiao, Hui and Xie, Hao and Wang, Haoqian and Fang, Lu and others},
journal={Nature Methods},
volume={18},
number={11},
pages={1395--1400},
year={2021},
publisher={Nature Publishing Group}
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
File details
Details for the file deepcad-1.3.0.tar.gz.
File metadata
- Download URL: deepcad-1.3.0.tar.gz
- Upload date:
- Size: 5.3 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.1 CPython/3.8.16
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
b7f0efc3802ce8731b71b181d503175c500d5062af40fae8ce1f6ef9c00a0b1a
|
|
| MD5 |
1f56bc0c178d92cc37c2f0f244c74040
|
|
| BLAKE2b-256 |
79c556c361ccdb822b8fda27f7e50106eb7eec1c697e0c8b32e09fc7a49a9364
|