A framework to train the activation functions of a neural network
Project description
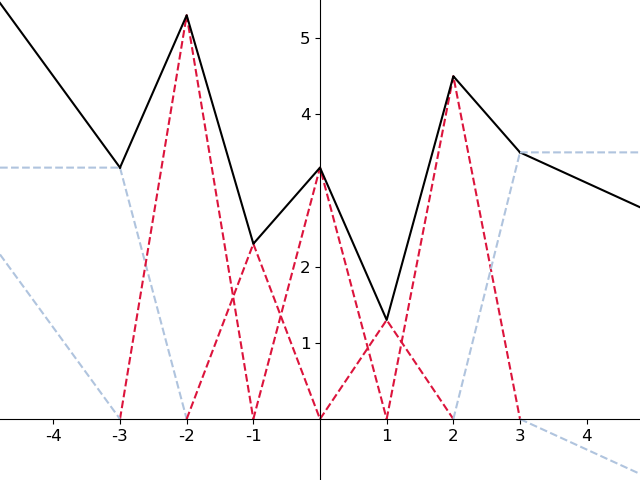

DeepSplines is a framework to train the activation functions of a neural network.
The aim of this repository is to:
Facilitate the reproduction of the results reported in the research papers:
Enable a seamless integration of deep spline activation functions in a custom neural network.
The proposed scheme is based on the theoretical work of M.Unser.
Requirements
python >= 3.7
numpy >= 1.10
pytorch >= 1.5.1
torchvision >= 0.2.2
matplotlib >= 3.3.1
(optional) CUDA
The code was developed and tested on a x86_64 Linux system.
Installation
To install the package, we first create an environment with python 3.7 (or greater):
>> conda create -y -n deepsplines python=3.7
>> source activate deepsplinesQuick Install
DeepSplines is available on Pypi. Therefore, you can install the package via the command:
>> pip install deepsplinesFor NVIDIA GPU compatibility, you need to additionally install cudatoolkit
(via conda install -c anaconda cudatoolkit)
Developper Install
It is also possible to install DeepSplines from the source for developpers:
>> git clone https://github.com/joaquimcampos/DeepSplines
>> cd <repository_dir>/
>> pip install -e .Usage
Here we show an example on how to adapt the PyTorch CIFAR-10 tutorial to use DeepBSpline activations.
from deepsplines.ds_modules import dsnn
class DSNet(dsnn.DSModule):
def __init__(self):
super().__init__()
self.conv_ds = nn.ModuleList()
self.fc_ds = nn.ModuleList()
# deepspline parameters
opt_params = {
'size': 51,
'range_': 4,
'init': 'leaky_relu',
'save_memory': False
}
# convolutional layer with 6 output channels
self.conv1 = nn.Conv2d(3, 6, 5)
self.conv_ds.append(dsnn.DeepBSpline('conv', 6, **opt_params))
self.pool = nn.MaxPool2d(2, 2)
self.conv2 = nn.Conv2d(6, 16, 5)
self.conv_ds.append(dsnn.DeepBSpline('conv', 16, **opt_params))
# fully-connected layer with 120 output units
self.fc1 = nn.Linear(16 * 5 * 5, 120)
self.fc_ds.append(dsnn.DeepBSpline('fc', 120, **opt_params))
self.fc2 = nn.Linear(120, 84)
self.fc_ds.append(dsnn.DeepBSpline('fc', 84, **opt_params))
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
x = self.pool(self.conv_ds[0](self.conv1(x)))
x = self.pool(self.conv_ds[1](self.conv2(x)))
x = torch.flatten(x, 1) # flatten all dimensions except batch
x = self.fc_ds[0](self.fc1(x))
x = self.fc_ds[1](self.fc2(x))
x = self.fc3(x)
return x
dsnet = DSNet()
dsnet.to(device)
main_optimizer = optim.SGD(dsnet.parameters_no_deepspline(),
lr=0.001,
momentum=0.9)
aux_optimizer = optim.Adam(dsnet.parameters_deepspline())
lmbda = 1e-4 # regularization weight
lipschitz = False # lipschitz control
for epoch in range(2):
for i, data in enumerate(trainloader):
# get the inputs; data is a list of [inputs, labels]
inputs, labels = data[0].to(device), data[1].to(device)
# zero the parameter gradients
main_optimizer.zero_grad()
aux_optimizer.zero_grad()
outputs = dsnet(inputs)
loss = criterion(outputs, labels)
# add regularization loss
if lipschitz is True:
loss = loss + lmbda * dsnet.BV2()
else:
loss = loss + lmbda * dsnet.TV2()
loss.backward()
main_optimizer.step()
aux_optimizer.step()For full details, please consult scripts/deepsplines_tutorial.py.
Reproducing results
To reproduce the results shown in the research papers [Bohra-Campos2020] and [Aziznejad2020] one can run the following scripts:
>> ./scripts/run_resnet32_cifar.py
>> ./scripts/run_nin_cifar.py
>> ./scripts/run_twoDnet.pyTo see the running options, please add --help to the commands above.
Developers
DeepSplines is developed by the Biomedical Imaging Group, École Polytéchnique Fédérale de Lausanne, Switzerland.
For citing this package, please see: http://doi.org/10.5281/zenodo.5156042
Original authors:
Joaquim Campos (joaquimcampos15@hotmail.com)
Pakshal Bohra (pakshal.bohra@epfl.ch)
Contributors:
Harshit Gupta
References
Bohra, J. Campos, H. Gupta, S. Aziznejad, M. Unser, “Learning Activation Functions in Deep (Spline) Neural Networks,” IEEE Open Journal of Signal Processing, vol. 1, pp.295-309, November 19, 2020.
Aziznejad, H. Gupta, J. Campos, M. Unser, “Deep Neural Networks with Trainable Activations and Controlled Lipschitz Constant,” IEEE Transactions on Signal Processing, vol. 68, pp. 4688-4699, August 10, 2020.
License
The code is released under the terms of the MIT License
Acknowledgements
This work was supported in part by the Swiss National Science Foundation under Grant 200020_184646 / 1 and in part by the European Research Council (ERC) under Grant 692726-GlobalBioIm.
MIT License
Copyright (c) 2021 Joaquim Campos, Pakshal Bohra
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the “Software”), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file deepsplines-1.0.0.tar.gz.
File metadata
- Download URL: deepsplines-1.0.0.tar.gz
- Upload date:
- Size: 58.6 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.2 importlib_metadata/4.6.3 pkginfo/1.7.1 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.0 CPython/3.7.11
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
2ec8703d4d31b5392af4f26af4ce46d837c5cff38cd09a82d8ea601f75429f79
|
|
| MD5 |
7712d3a808df629aa9e4f82c72b4c720
|
|
| BLAKE2b-256 |
e831147bafb18a3ed7b2aade192bd601369e0a10dd801ae631cefce8e6b4356b
|
File details
Details for the file deepsplines-1.0.0-py3-none-any.whl.
File metadata
- Download URL: deepsplines-1.0.0-py3-none-any.whl
- Upload date:
- Size: 79.2 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.2 importlib_metadata/4.6.3 pkginfo/1.7.1 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.0 CPython/3.7.11
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
0aba1fd72b613b396b0f7ed658ce410b789fc5004b993a8f0e62a4048786287c
|
|
| MD5 |
905fe238fb99ae0910639b0952b0b1c0
|
|
| BLAKE2b-256 |
a4d7c8129172b78659541f42a8e52e3a4d65cbf2cfb487970aabb24b9ae11b09
|











