densMAP
Project description
densMAP
This software package contains an implementation of density-preserving data visualization tool densMAP, which augments the UMAP algorithm (based on v0.3.10). Some of the following instructions are adapted from the UMAP repository.
Installation
densMAP shares the same dependencies as UMAP, including:
- numpy
- scipy
- scikit-learn
- numba==0.48.0
Our code currently does not support the latest version of numba (0.49.0).
Install Options
PyPI installation of densMAP can be performed as:
pip install densmap-learn
For a manual install, first download this package:
wget https://github.com/hhcho/densvis/archive/master.zip
unzip densvis-master.zip
rm densvis-master.zip
cd densvis-master/densmap/
Install the requirements:
sudo pip install -r requirements.txt
or
conda install scikit-learn numba
Finally, install the package:
python setup.py install
Usage
Like UMAP, the densMAP package inherits from sklearn classes, and thus drops in neatly next to other sklearn transformers with an identical calling API.
import densmap
from sklearn.datasets import fetch_openml
from sklearn.utils import resample
digits = fetch_openml(name='mnist_784')
subsample, subsample_labels = resample(digits.data, digits.target, n_samples=7000,
stratify=digits.target, random_state=1)
embedding, ro, re = densmap.densMAP().fit_transform(subsample)
Input arguments
There are a number of parameters that can be set for the densMAP class; the major ones inherited from UMAP are:
-
n_neighbors: This determines the number of neighboring points used in local approximations of manifold structure. Larger values will result in more global structure being preserved at the loss of detailed local structure. In general this parameter should often be in the range 5 to 50; we set a default of 30. -
min_dist: This controls how tightly the embedding is allowed compress points together. Larger values ensure embedded points are more evenly distributed, while smaller values allow the algorithm to optimise more accurately with regard to local structure. Sensible values are in the range 0.001 to 0.5, with 0.1 being a reasonable default. -
metric: This determines the choice of metric used to measure distance in the input space. A wide variety of metrics are already coded, and a user defined function can be passed as long as it has been JITd by numba.
The additional parameters specific to densMAP are:
-
dens_frac: This determines the fraction of iterations that will include the density-preservation term of the gradient (float, between 0 and 1); default 0.3. -
dens_lambda: This determines the weight of the density-preservation objective. See the original paper for the effect this parameter has when changed (float, non-negative); default 2.0. -
final_dens: When this flag isTrue, the code returns, in addition to the embedding, the local radii for the original dataset and for the embedding. IfFalse, only the embedding is returned (bool); defaultTrue.
Other parameters that can be set include:
-
ndim: Dimensions of the embedding (int); default 2. -
n_epochs: Number of epochs to run the algorithm (int); default 750. -
var_shift: Regularization term added to the variance of embedding local radius for stability (float, non-negative); default 0.1.
Output arguments
If final_dens is True, returns (embedding, ro, re), where:
-
embedding: a (number of data points)-by-ndimsnumpy array containing the embedding coordinates -
ro: a numpy array of length (number of data points) that contains the log local radius of the input data -
re: a numpy array of length (number of data points) that contains the log local radius of the embedded data
If final_dens is False, returns just embedding.
An example of making use of these options:
embedding, ro, re = densmap.densMAP(n_neighbors=25, n_epochs=500, dens_frac=0.3,
dens_lambda=0.5).fit_transform(data)
R wrapper
We use the reticulate library to provide compatibility with R as well with the
script densmap.R. Since reticulate runs Python code with an R wrapper, to use this
library you must have Python3 installed. The script will automatically install the
densmap-learn package via pip if it is not installed.
From then, within your R script, you can run
source("densmap.R")
# Assume `data` is an R dataframe, needs to be converted to a matrix
out <- densMAP(as.matrix(data))
The R function densMAP takes the same optional arguments listed in Input Arguments section
above with the same names and default values. So you can, for example, run:
out <- densMAP(as.matrix(data), n_neighbors=25, n_epochs=500, dens_frac=0.3, dens_lambda=0.5)
If final_dens is TRUE then out[[1]] will contain the embedding, out[[2]] will be the
log original local radii, and out[[3]] the log embedding local radii.
If final_dens is FALSE then out will be the embeddings itself.
From the command line
We also provide the file densmap.py which allows you to run densMAP from the terminal,
specifying the major options from above. Simply run:
python densmap.py -i [--input INPUT] -o [--outname OUTNAME] -f [--dens_frac DENS_FRAC]
-l [--dens_lambda DENS_LAMBDA] -s [--var_shift VAR_SHIFT] -d [--ndim NDIM]
-n [--n-epochs N-EPOCHS] -k [--n-nei N-NEIGHBORS] [--final_dens/--no_final_dens FINAL_DENS]
where within the square braces are the long-form flag and the capitalized text corresponds to the parameters above. For example:
python densmap.py -i data.txt -o out -f .3 -k 25
and
python densmap.py --input data.txt --outname out --dens_frac .3 --n-nei 25
both run densMAP on input file data.txt to produce output files out_emb.txt
and out_dens.txt, using dens_frac=0.3 and n_neighbors=25.
The input file is parsed using numpy’s loadtxt function if it is a .txt file; another option is to provide a .pkl file.
We assume that the first dimension (row index) iterates over the data instances, and the second dimension (column index) iterates over the features.
The output files include:
out_emb.txta TSV file containing the embedding coordinatesof the data, andout_dens.txta (number of data points)-by-2 TSV file containing in the first column the log local radii in the original data and in the second column the log local radii in the embedding.
Example data and script
We have included the file trial_densmap.py which allows you to run an example straight out of
the box.
Run:
python trial_densmap.py
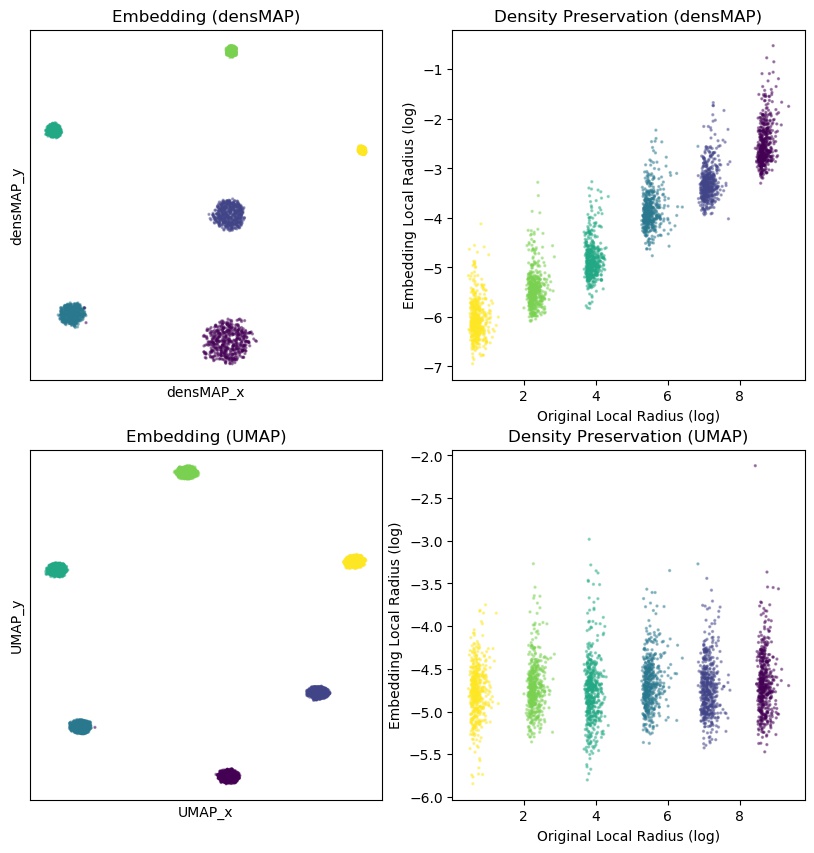
The code will load a dataset that contains a mixture of six Gaussian point clouds
with increasing variance and
will run both densMAP and UMAP on the dataset with default parameters and plot the embeddings
(if you have matplotlib installed),
and alignment of the local radius in each case. It will also save the embeddings in {umap,densmap}_trial_emb.txt,
the local radii in {umap,densmap}_trial_dens.txt, and the plot in densmap_trial_fig.png.
The plot will look like:
References
Our densMAP algorithm is described in:
Ashwin Narayan, Bonnie Berger*, and Hyunghoon Cho*. "Density-Preserving Data Visualization Unveils Dynamic Patterns of Single-Cell Transcriptomic Variability", bioRxiv, 2020.
Original UMAP algorithm is described in:
Leland McInnes, John Healy, and James Melville. "UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction", arXiv, 1802.03426, 2018.
License
This package is licensed under the MIT license.
Contact for questions
Ashwin Narayan, ashwinn@mit.edu
Hoon Cho, hhcho@broadinstitute.org
Additionally, some questions regarding the UMAP-specific aspects of this software may be answered by browsing the UMAP documentation at Read the Docs, which includes an FAQ.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file densmap-learn-0.2.2.tar.gz.
File metadata
- Download URL: densmap-learn-0.2.2.tar.gz
- Upload date:
- Size: 361.9 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.2.0 pkginfo/1.6.1 requests/2.25.0 setuptools/50.3.2 requests-toolbelt/0.9.1 tqdm/4.51.0 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
004f66f0fbddfd5f36cdde29abdb0e59b83fc9d15ef9373114194fc3430a0800
|
|
| MD5 |
6631e689fd05b2fba26ead55475d3964
|
|
| BLAKE2b-256 |
b9f65671fa4ac7ea1f911920f7e7d793e23c0aee100104adc5c92cb4279fbeb0
|
File details
Details for the file densmap_learn-0.2.2-py3-none-any.whl.
File metadata
- Download URL: densmap_learn-0.2.2-py3-none-any.whl
- Upload date:
- Size: 680.4 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.2.0 pkginfo/1.6.1 requests/2.25.0 setuptools/50.3.2 requests-toolbelt/0.9.1 tqdm/4.51.0 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
e55a9001d0466651abb29b4f1ac0f810b9fbb05e70d86af39235158cb19cdd89
|
|
| MD5 |
7ab87b0edb4b16f3ae83f01a9659fb97
|
|
| BLAKE2b-256 |
95435e471ff639e28e1e76ebfcbf211ab3c90542367f138383ea62052edc5c8c
|











