An accompanying package for the paper, Deep Goal-Oriented Clustering
Project description
Deep Goal-Oriented Clustering
This is the depository for the paper, Deep Goal-Oriented Clustering (DGC). This depository contains code to replicate the CIFAR 100-20 experiment detailed in the paper. Here we give a brief description of DGC.
DGC is built upon VAE, and uses similar variational techniques to maximize a variation lower bound of the data log-likelihood. A (deep) variational method can be efficiently summarized in terms of its generative & infernece steps, which we describe next.
Generative process for DGC
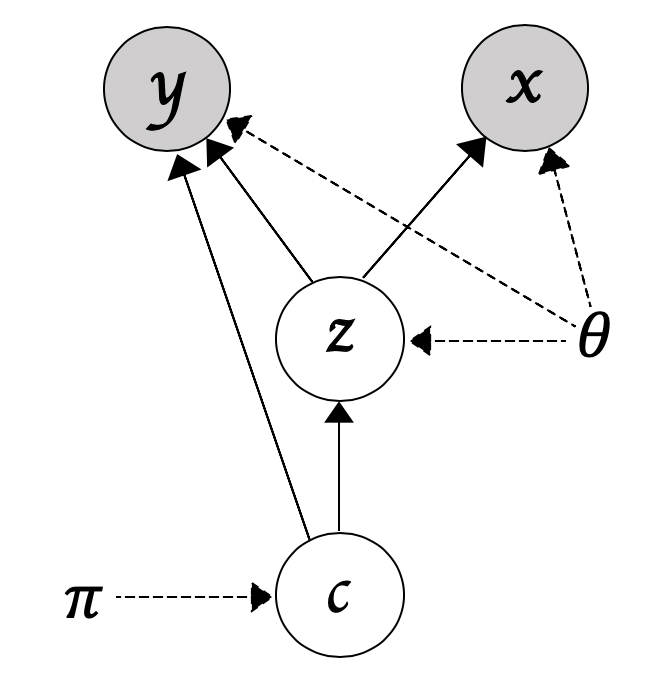
Let x,y,z and c denote the input data, the side-information, the latent code, and the index for a given Gaussian mixture component, we then have
p(x,y,z,c) = p(y|z,c)p(x|z)p(z|c)p(c)
In words, we first sample a component index from p(c), sample the latent code z from p(z|c), and then we reconstruct the input x and predict for the side-information y (see the figure below for a figurative illustration).
Inference for DGC
For the variational lower bound of DGC, please refer to Eq. 2 in the main paper. In a nutshell, we want to maximize the log-likelihood by maximizing its variational lower bound.
Test the model on Pacman
To run the model on the Pacman dataset, first install the package
pip install dgc
After the installation, simply follow the following
# Test model on the sythetic dataset Pacman
from dgc import load_sample_datasets
from dgc import dgc
dataset = 'pacman'
task_name = 'regression'
batch_size = 128
learning_rate = 0.01
epochs = 50
trainloader, testloader, _ = load_sample_datasets(batch_size,dataset)
model = dgc(input_dim=2, y_dim = 1, z_dim=10, n_centroids=2, task = task_name, binary=True,
encodeLayer=[128,256,256], decodeLayer=[256,256,128])
model.fit(trainloader, testloader, lr=learning_rate, num_epochs=epochs,
anneal=True, direct_predict_prob=False)
Test the model on your dataset
To run DGC on your own dataset, you will need to have the following files (all of which are assumed to be numpy arrays)
- train_features.npy: this contains the training features
- train_side_info.npy: this contains the training side-information (can be either discrete or continous)
- train_cluster_labels.npy: this contains the training clustering labels
- test_features.npy: this contains the test features
- test_side_info.npy: this contains the test side-information (can be either discrete or continous)
- test_cluster_labels.npy: this contains the test clustering labels
You can create your own dataloader or passing the files into the built-in loader function
from dgc import form_dataloaders
from dgc import dgc
batch_size = 128 # whatever you want
task_name = 'regression' # or classification
batch_size = 128
learning_rate = 0.01
epochs = 50
input_dim = 2
y_dim = 1,
z_dim = 10,
n_centroids = 10
binary = True
encoder_size = [128,256,256]
decoder_size = [256,256,128]
learning_rate = 0.01
num_epochs = 10
anneal = True
direct_predict_prob = False
save_model_path = './sample_model.pt'
train_data = [train_features,train_side_info,train_cluster_labels] # order matters here
test_data = [test_features,test_side_info,test_cluster_labels]
trainloader,testloader,_ = form_dataloaders(batch_size,train_data,test_data)
model = dgc(input_dim=input_dim, y_dim = y_dim, z_dim=z_dim, n_centroids=n_centroids, task = task_name, binary=binary,
encodeLayer=encoder_size, decodeLayer=decoder_size)
model.fit(trainloader, testloader, lr=learning_rate, num_epochs=epochs
anneal=anneal, direct_predict_prob=direct_predict_prob)
# If you want to save the model
model.save_model(save_model_path)
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file dgc-0.1.3.tar.gz.
File metadata
- Download URL: dgc-0.1.3.tar.gz
- Upload date:
- Size: 24.6 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.8.0 pkginfo/1.8.2 readme-renderer/32.0 requests/2.26.0 requests-toolbelt/0.9.1 urllib3/1.26.7 tqdm/4.62.3 importlib-metadata/4.8.1 keyring/23.1.0 rfc3986/2.0.0 colorama/0.4.4 CPython/3.9.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
32072255469f883a0ed65b5fda15739f94a689a0442567022bd9984e9bc32bc8
|
|
| MD5 |
84e5cc4bc10241364c28324a3a551647
|
|
| BLAKE2b-256 |
4f34703259f7689541ed8d65e8e59c769c4bc9f390afb22229c3884511513dd6
|
File details
Details for the file dgc-0.1.3-py3-none-any.whl.
File metadata
- Download URL: dgc-0.1.3-py3-none-any.whl
- Upload date:
- Size: 26.6 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.8.0 pkginfo/1.8.2 readme-renderer/32.0 requests/2.26.0 requests-toolbelt/0.9.1 urllib3/1.26.7 tqdm/4.62.3 importlib-metadata/4.8.1 keyring/23.1.0 rfc3986/2.0.0 colorama/0.4.4 CPython/3.9.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
54ba3451bc3159fc96a237e22afac152dc707182fcc05d275155282e96822b2c
|
|
| MD5 |
9c01708c72e7e5352bc225adc690c2ad
|
|
| BLAKE2b-256 |
69aa7be09c1f16dc937cfb0176463b02bb70476dc8b5711749c63ddc559e34cb
|