Infer Functional Associations using Variational Autoencoders on -Omics data.
Project description
 FAVA: Functional Associations using Variational Autoencoders
FAVA: Functional Associations using Variational Autoencoders
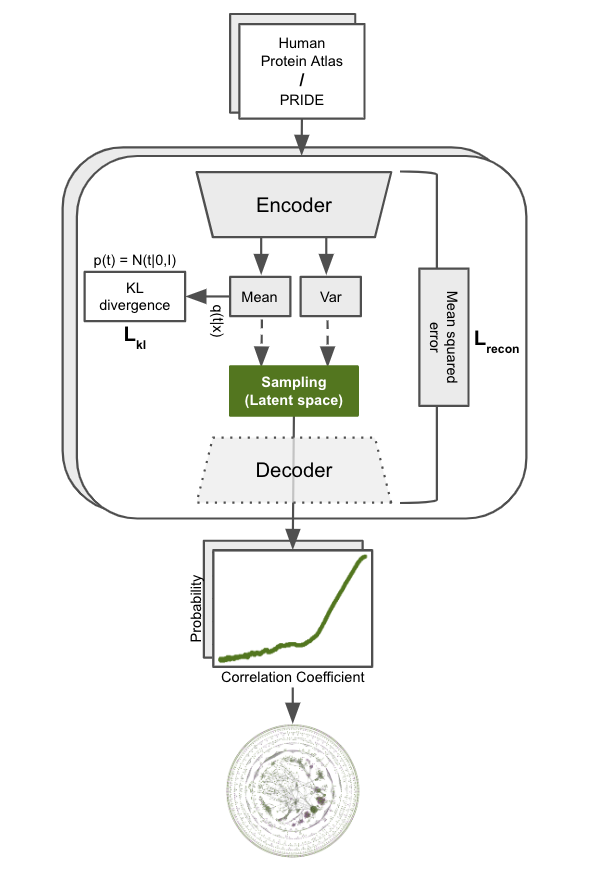
FAVA is a method used to construct protein networks based on omics data such as single-cell RNA sequencing (scRNA-seq) and proteomics. Existing protein networks are often biased towards well-studied proteins, limiting their ability to reveal functions of understudied proteins. FAVA addresses this issue by leveraging omics data that are not influenced by literature bias. Read the documentation.
Data availability
Installation:
pip install favapy
favapy as Python library
Read the jupyter-notebook: How_to_use_favapy_in_a_notebook
favapy supports both AnnData objects and count/abundance matrices.
Command line interface
Run favapy from the command line as follows:
favapy <path-to-data-file> <path-to-save-output>
Optional parameters:
-t Type of input data ('tsv' or 'csv'). Default value = 'tsv'.
-n The number of interactions in the output file (with both directions, proteinA-proteinB and proteinB-proteinA). Default value = 100000.
-cor Type of correlation method ('pearson' or 'spearman'). Default value = 'pearson'
-c The cut-off on the Correlation scores.The scores can range from 1 (high correlation) to -1 (high anti-correlation). This option overwrites the number of interactions. Default value = None.
-d The dimensions of the intermediate\hidden layer. Default value depends on the input size.
-l The dimensions of the latent space. Default value depends on the size of the hidden layer.
-e The number of epochs. Default value = 50.
-b The batch size. Default value = 32.
If FAVA is useful for your research, consider citing FAVA BiorXiv.
Other Relevant publications:
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file favapy-1.0.1.tar.gz.
File metadata
- Download URL: favapy-1.0.1.tar.gz
- Upload date:
- Size: 10.2 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.0 CPython/3.9.19
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
b72376dc851eb4bfd6be10a60427bc0f7fac1378dbacde87a3983b9722961bda
|
|
| MD5 |
b6df6d719da51116fb2190b1dadca064
|
|
| BLAKE2b-256 |
f0ff5b35f6fb0431df6c6630d49572ab5ce0bc10b850b02c6ef19d0bc2091ec9
|
File details
Details for the file favapy-1.0.1-py3-none-any.whl.
File metadata
- Download URL: favapy-1.0.1-py3-none-any.whl
- Upload date:
- Size: 8.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.0 CPython/3.9.19
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
35099212c59ebf768557627a66f2cdb8096a7b652b34d5656d824aefe4dc94da
|
|
| MD5 |
4e7a126778d0e81b970c43ee97613325
|
|
| BLAKE2b-256 |
70e47a06208e81b70e16c6717804b8cb313eaa66a63dc70cbedd16e07e8b64d8
|