Core bioimage-analysis library and tasks of the Fractal analytics platform
Project description
Fractal Core Tasks
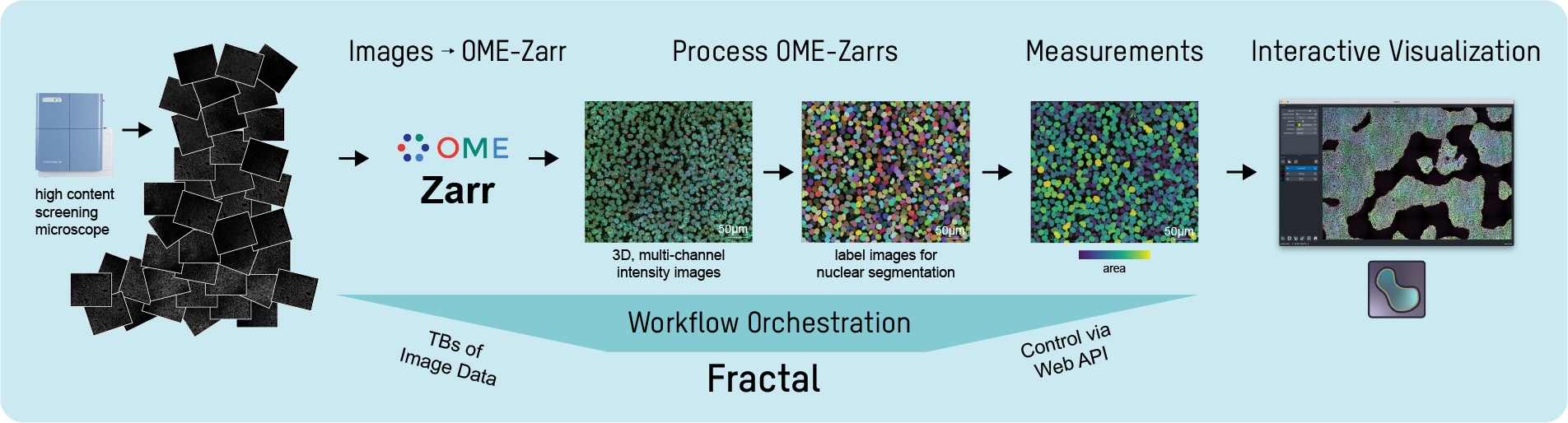
Fractal is a framework to process high-content imaging data at scale and prepare it for interactive visualization.
Fractal provides distributed workflows that convert TBs of image data into OME-Zarr files. The platform then processes the 3D image data by applying tasks like illumination correction, maximum intensity projection, 3D segmentation using cellpose and measurements using napari workflows. The pyramidal OME-Zarr files enable interactive visualization in the napari viewer.
This is the core-tasks repository, containing the python tasks that parse Yokogawa CV7000 images into OME-Zarr and process OME-Zarr files. Find more information about Fractal in general and the other repositories at the Fractal home page.
Documentation
See https://fractal-analytics-platform.github.io/fractal-tasks-core
Available tasks
Currently, the following tasks are available:
- Create Zarr Structure: Task to generate the zarr structure based on Yokogawa metadata files
- Yokogawa to Zarr: Parses the Yokogawa CV7000 image data and saves it to the Zarr file
- Illumination Correction: Applies an illumination correction based on a flatfield image & subtracts a background from the image.
- Image Labeling (& Image Labeling Whole Well): Applies a cellpose network to the image of a single ROI or the whole well. cellpose parameters can be tuned for optimal performance.
- Maximum Intensity Projection: Creates a maximum intensity projection of the whole plate.
- Measurement: Make some standard measurements (intensity & morphology) using napari workflows, saving results to AnnData tables.
Some additional tasks are currently being worked on and some older tasks are still present in the fractal_tasks_core folder.
Contributors and license
Unless otherwise stated in each individual module, all Fractal components are released according to a BSD 3-Clause License, and Copyright is with Friedrich Miescher Institute for Biomedical Research and University of Zurich.
Fractal was conceived in the Liberali Lab at the Friedrich Miescher Institute for Biomedical Research and in the Pelkmans Lab at the University of Zurich (both in Switzerland). The project lead is with @gusqgm & @jluethi. The core development is done under contract by eXact lab S.r.l..
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for fractal_tasks_core-0.12.1.tar.gz
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 5e69015db9cc4daa380f837cd3a6559d385cc42b490993cce86fc662a6a744d2 |
|
| MD5 | 5f709dd1861d35f276e1a39f70bbbbe8 |
|
| BLAKE2b-256 | 335f311017ffdf2d1996d2e775e1e02a2fb782751d1ef3adc11d503d97c140ed |
Hashes for fractal_tasks_core-0.12.1-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 77286a9baa2a2052039d417b4defbe260b337fbdb3e0b26b51a095b439eb7117 |
|
| MD5 | d9726fdebad90887f81a3796def5592a |
|
| BLAKE2b-256 | ee0f9802d59ae5694dfc25b8cd965736433717bda02842fd5c20cf502aaae167 |