Predicting FRET with accessible-contact volumes
Project description

FRETraj is a Python module for calculating multiple accessible-contact volumes (multi-ACV) and predicting FRET efficiencies. It provides an interface to the LabelLib library to simulate fluorophore dynamics. The package features a user-friendly PyMOL plugin1 which can be used to explore different labeling positions when designing FRET experiments. In an AV simulation the fluorophore distribution is estimated by a shortest path search (Djikstra algorithm) using a coarse-grained dye probe. FRETraj further implements a Python-only version of the geometrical clash search used in LabelLib. This should facilitate prototyping of new features for the ACV algorithm.
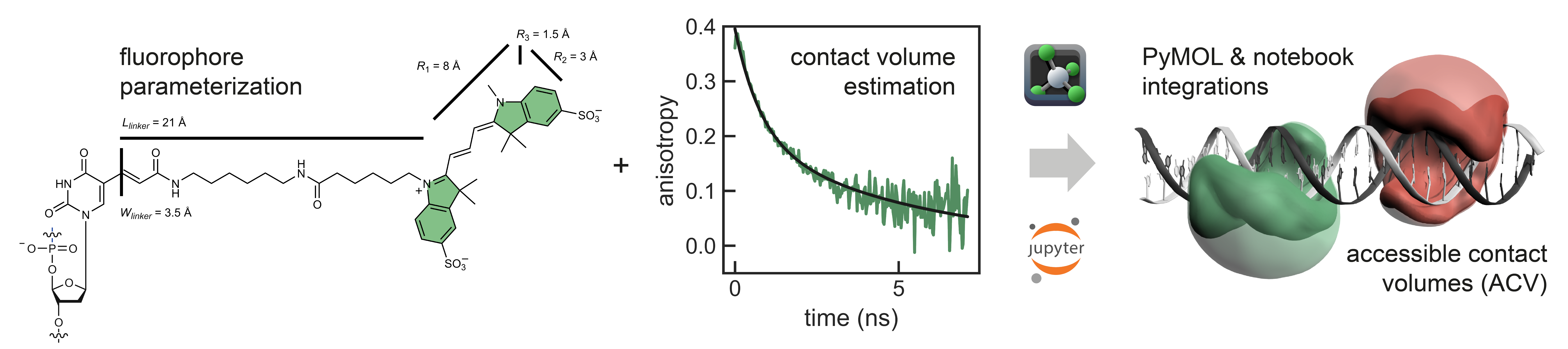
A recent addition to the original AV model (Kalinin et al. Nat. Methods, 2012) is the so-called contact volume (Steffen et. al. PCCP 2016). Here, the accessible volume is split into a free volume (FV, transparent) where the dye is freely diffusing and a contact volume (CV, opaque) where the dye stacks to the biomolecular surface. Time-resolved anisotropy experiments suggest that certain fluorophores, among those the commonly used cyanines Cy3 and Cy5, are particularly prone to interact with both proteins and nucleic acids. The contact volume accounts for this effect by reweighting the point-cloud. By choosing different experimental weights for the free and contact component the AV dye model is refined, making in silico FRET predictions more reliable.
Installation
Follow the instructions for your platform here
References
If you use FRETraj in your work please refer to the following paper:
Additional readings
- S. Kalinin, T. Peulen, C.A.M. Seidel et al. Nat. Methods, 2012, 9, 1218-1225.
- T. Eilert, M. Beckers, F. Drechsler, J. Michaelis, Comput. Phys. Commun., 2017, 219, 377–389.
- M. Dimura, T. Peulen, C.A.M. Seidel et al. Curr. Opin. Struct. Biol. 2016, 40, 163-185.
- M. Dimura, T. Peulen, C.A.M Seidel et al. Nat. Commun. 2020, 11, 5394.
1 PyMOL is a trademark of Schrödinger, LLC.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for fretraj-0.1.5a0-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 15c58685403076079ffa11bb2a2228da3bb458735c45b058bdee0625915bff0e |
|
| MD5 | b6f7000adaf5b07e042a54f7453493a0 |
|
| BLAKE2b-256 | c53ab53a6b1069178afdc86cd6eb9898eff0bfdbff4caec2edd82cfd46376f89 |

















