Performant subgraph isomorphism
Project description
Grand Isomorphisms
Subgraph isomorphism is a resource-heavy (but branch-parallelizable) algorithm that is hugely impactful for large graph analysis. SotA algorithms for this (Ullmann, VF2, BB-Graph) are heavily RAM-bound, but this is due to a large number of small processes each of which hold a small portion of a traversal tree in memory.
Grand-Iso is a subgraph isomorphism algorithm that exchanges this resource-limitation for a parallelizable partial-match queue structure.
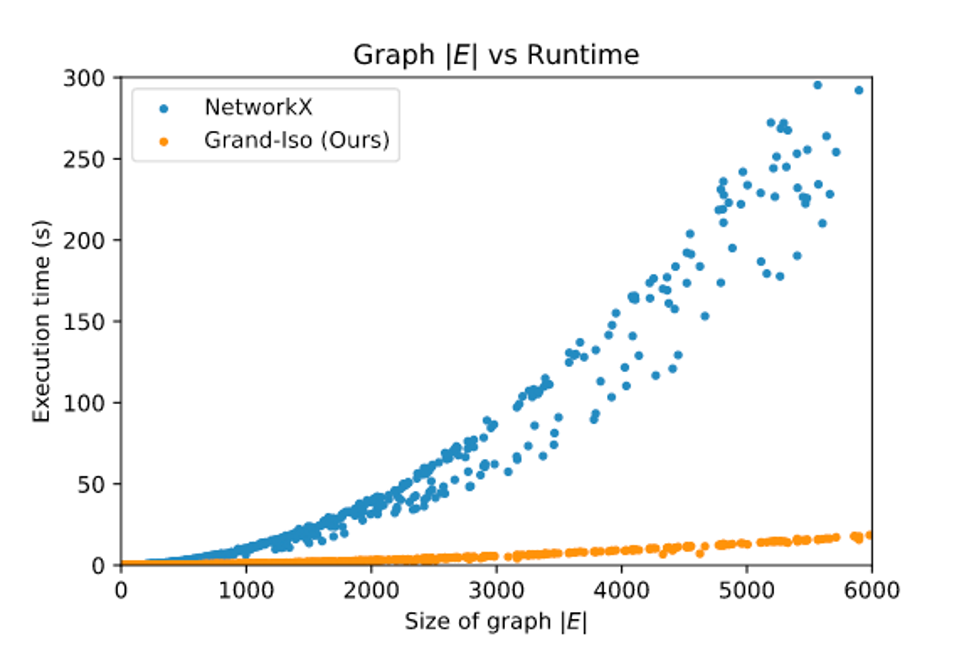
It performs favorably compared to other pure-python (and even some non-pure-python!) implementations:
Example Usage
from grandiso import find_motifs
import networkx as nx
host = nx.fast_gnp_random_graph(10, 0.5)
motif = nx.Graph()
motif.add_edge("A", "B")
motif.add_edge("B", "C")
motif.add_edge("C", "D")
motif.add_edge("D", "A")
len(find_motifs(motif, host))
Directed graph support:
from grandiso import find_motifs
import networkx as nx
host = nx.fast_gnp_random_graph(10, 0.5, directed=True)
motif = nx.DiGraph()
motif.add_edge("A", "B")
motif.add_edge("B", "C")
motif.add_edge("C", "D")
motif.add_edge("D", "A")
len(find_motifs(motif, host))
Counts-only
For very large graphs, you may use a good chunk of RAM not only on the queue of hypotheses, but also on the list of results. If all you care about is the NUMBER of results, you should pass count_only=True to the find_motifs function. This will dramatically reduce your RAM overhead on higher-count queries.
All Arguments to find_motifs
| Argument | Type | Default | Description |
|---|---|---|---|
motif |
nx.Graph |
The motif to search for. Either a nx.Graph or nx.DiGraph. You can also pass something networkx.Graph-flavored, like a grand.Graph. |
|
host |
nx.Graph |
The "haystack" graph to search within. Either a nx.Graph or nx.DiGraph. You can also pass something networkx.Graph-flavored, like a grand.Graph. |
|
interestingness |
dict |
None |
A lookup dictionary that assigns a floating number between 0 and 1 to each node in the motif graph. Nodes with values closest to 1 will be preferred when the candidate-mapper advances the exploration front. If this doesn't make sense to you, you can ignore this argument entirely. If none is provided, this will default to a uniform interestness metric where all nodes are considered equally. |
count_only |
bool |
False |
Whether to return only a count of motifs, rather than a list of mappings. If you set this to True, you will get back an integer result, not a list of dicts. |
directed |
bool |
None |
Whether to enforce a directed/undirected search. True means enforce directivity; False means enforce undirected search. The default (None) will guess based upon your motif and host. |
profile |
bool |
False |
Whether to slow down execution but give you a better idea of where your RAM usage is going. This is better ignored unless you're debugging something particularly nuanced. |
isomorphisms_only |
bool |
False |
Whether to search only for isomorphisms. In other words, whether to search for edges that exist in the node-induced subgraph. |
hints |
List[Dict[Hashable, Hashable]] |
None |
A list of valid candidate mappings to use as the starting seeds for new maps. See Using Hints, below. |
limit |
int |
None |
An optional integer limit of results to return. If the limit is reached, the search will return early. |
Using Hints
GrandIso optionally accepts an argument hints which is a list of valid partial mappings to use to seed the search. For example, in this code:
host = nx.DiGraph()
nx.add_path(host, ["A", "B", "C", "A"])
motif = nx.DiGraph()
nx.add_path(motif, ["a", "b", "c", "a"])
find_motifs(motif, host)
There are three valid mappings (because each of A, B, and C can map to a, b, or c).
We can declare that node A maps to node a or b like this:
host = nx.DiGraph()
nx.add_path(host, ["A", "B", "C", "A"])
motif = nx.DiGraph()
nx.add_path(motif, ["a", "b", "c", "a"])
find_motifs(
motif, host,
hints=[{"A": "a"}, {"A", "b"}]
)
Pseudocode for "Grand-Iso" algorithm
- Accept a motif M, and a host graph H.
- Create an empty list for result storage, R.
- Create an empty queue, Q.
- Preprocessing
- Identify the most "interesting" node in motif M, m1.
- Add to Q a set of mappings with a single node, with one map for all
nodes in H that satisfy the requirements of m1: degree, attributes, etc
- Motif Search
- "Pop" a backbone B from Q
- Identify as m1 the most interesting node in motif M that does not yet
have a mapping assigned in B.
- Identify all nodes that are valid mappings from the backbone to m1,
based upon degree, attributes, etc.
- If multiple nodes are valid candidates, add each new backbone to Q.
- Otherwise, when all nodes in M have a valid mapping in B to H, add
the mapping to the results set R.
- Continue while there are still backbones in Q.
- Reporting
- Return the set R to the user.
Hacking on this repo
Running Tests
coverage run --source=grandiso -m pytest
Citing
If this tool is helpful to your research, please consider citing it with:
# https://doi.org/10.1101/2020.06.08.140533
@article{matelsky_2020_dotmotif,
title={{DotMotif: An open-source tool for connectome subgraph isomorphism search and graph queries}},
url={http://dx.doi.org/10.1101/2020.06.08.140533},
DOI={10.1101/2020.06.08.140533},
publisher={Cold Spring Harbor Laboratory},
author={Matelsky, Jordan K. and Reilly, Elizabeth P. and Johnson, Erik C. and Stiso, Jennifer and Bassett, Danielle S. and Wester, Brock A. and Gray-Roncal, William},
year={2020},
month={Jun}
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
File details
Details for the file grandiso-1.1.0.tar.gz.
File metadata
- Download URL: grandiso-1.1.0.tar.gz
- Upload date:
- Size: 13.5 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.2.0 pkginfo/1.6.1 requests/2.23.0 setuptools/41.2.0 requests-toolbelt/0.9.1 tqdm/4.44.1 CPython/3.7.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
6ceae4ebac2a29a766fda112e268855e70a73e7361f2571cf4d2deb6090071d8
|
|
| MD5 |
be1e2cfeaf866a412bb2e86dd83acf4d
|
|
| BLAKE2b-256 |
a6b9f567101b375ffcf2dbd8bb2ec9128112a023b6638c95ab91019d93522205
|















