A visual graph analytics library for extracting, transforming, displaying, and sharing big graphs with end-to-end GPU acceleration
Project description
PyGraphistry: Leverage the power of graphs & GPUs to visualize, analyze, and scale your data
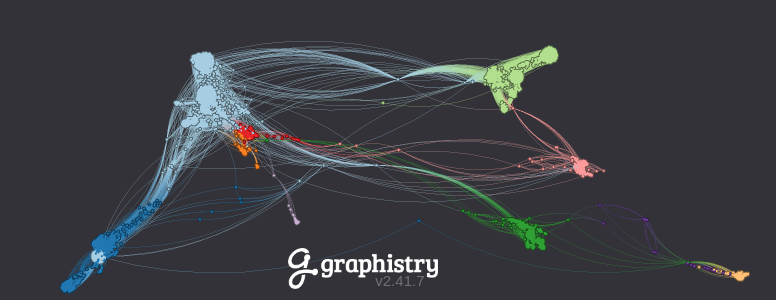 Demo: Interactive visualization of 80,000+ Facebook friendships (source data)
Demo: Interactive visualization of 80,000+ Facebook friendships (source data)
|
PyGraphistry is an open source Python library for data scientists and developers to leverage the power of graph visualization, analytics, AI, including with native GPU acceleration:
-
Python dataframe-native graph processing: Quickly ingest & prepare data in many formats, shapes, and scales as graphs. Use tools like Pandas, Spark, RAPIDS (GPU), and Apache Arrow.
-
Integrations: Connect to graph databases, data platforms, Python tools, and more.
Category Connector Tutorials Data Platforms, SQL & Logs Graph Databases Python Tools & Libraries -
Prototype locally and deploy remotely: Prototype from notebooks like Jupyter and Databricks using local CPUs & GPUs, and then power production dashboards & pipelines with Graphistry Hub and your own self-hosted servers.
-
Query graphs with GFQL: Use GFQL, the first dataframe-native graph query language, to ask relationship questions that are difficult for tabular tools and without requiring a database.
-
graphistry[ai]: Call streamlined graph ML & AI methods to benefit from clustering, UMAP embeddings, graph neural networks, automatic feature engineering, and more.
-
Visualize & explore large graphs: In just a few minutes, create stunning interactive visualizations with millions of edges and many point-and-click built-ins like drilldowns, timebars, and filtering. When ready, customize with Python, JavaScript, and REST APIs.
-
Columnar & GPU acceleration: CPU-mode ingestion and wrangling is fast due to native use of Apache Arrow and columnar analytics, and the optional RAPIDS-based GPU mode delivers 100X+ speedups.
From global 10 banks, manufacturers, news agencies, and government agencies, to startups, game companies, scientists, biotechs, and NGOs, many teams are tackling their graph workloads with Graphistry.
Gallery
The notebook demo gallery shares many more live visualizations, demos, and integration examples
Twitter Botnet |
Edit Wars on Wikipedia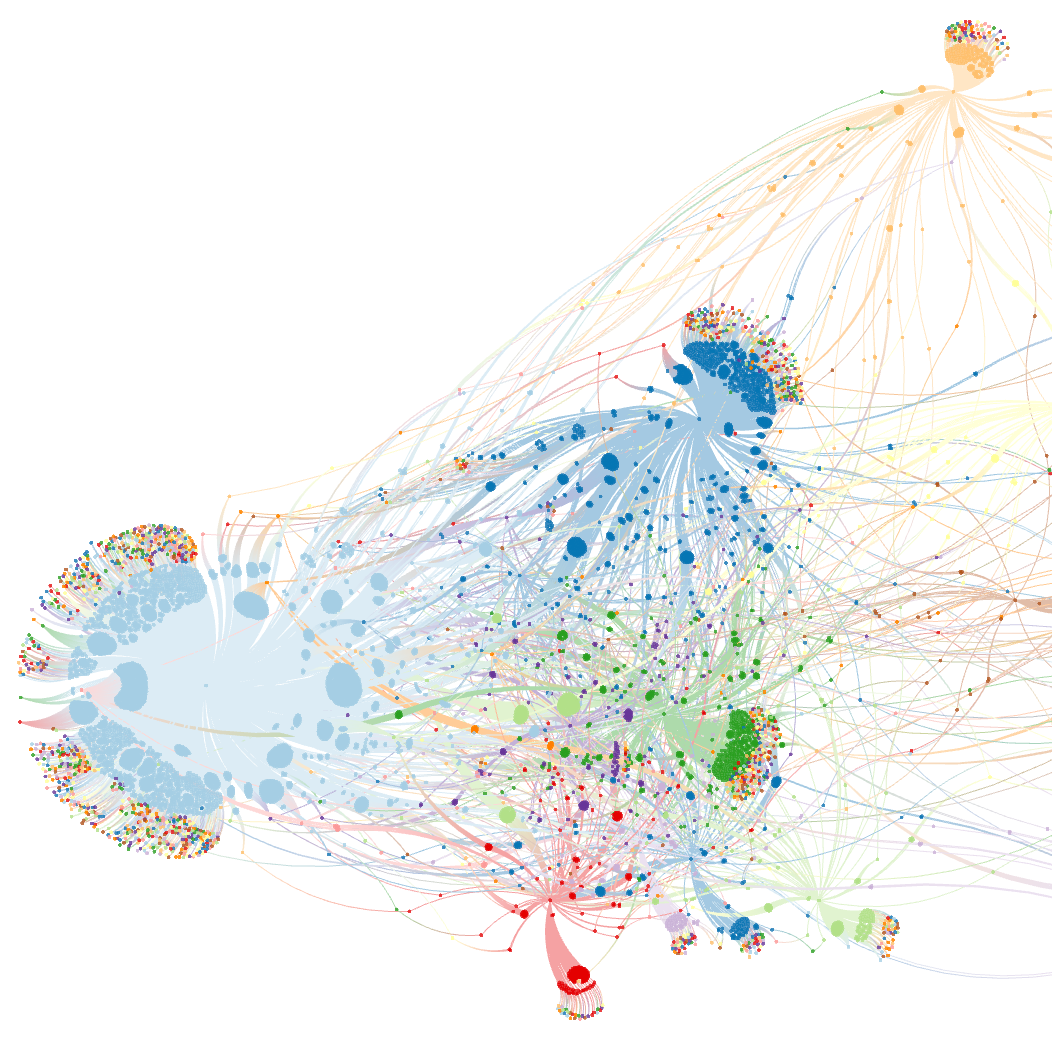 (data) (data) |
100,000 Bitcoin Transactions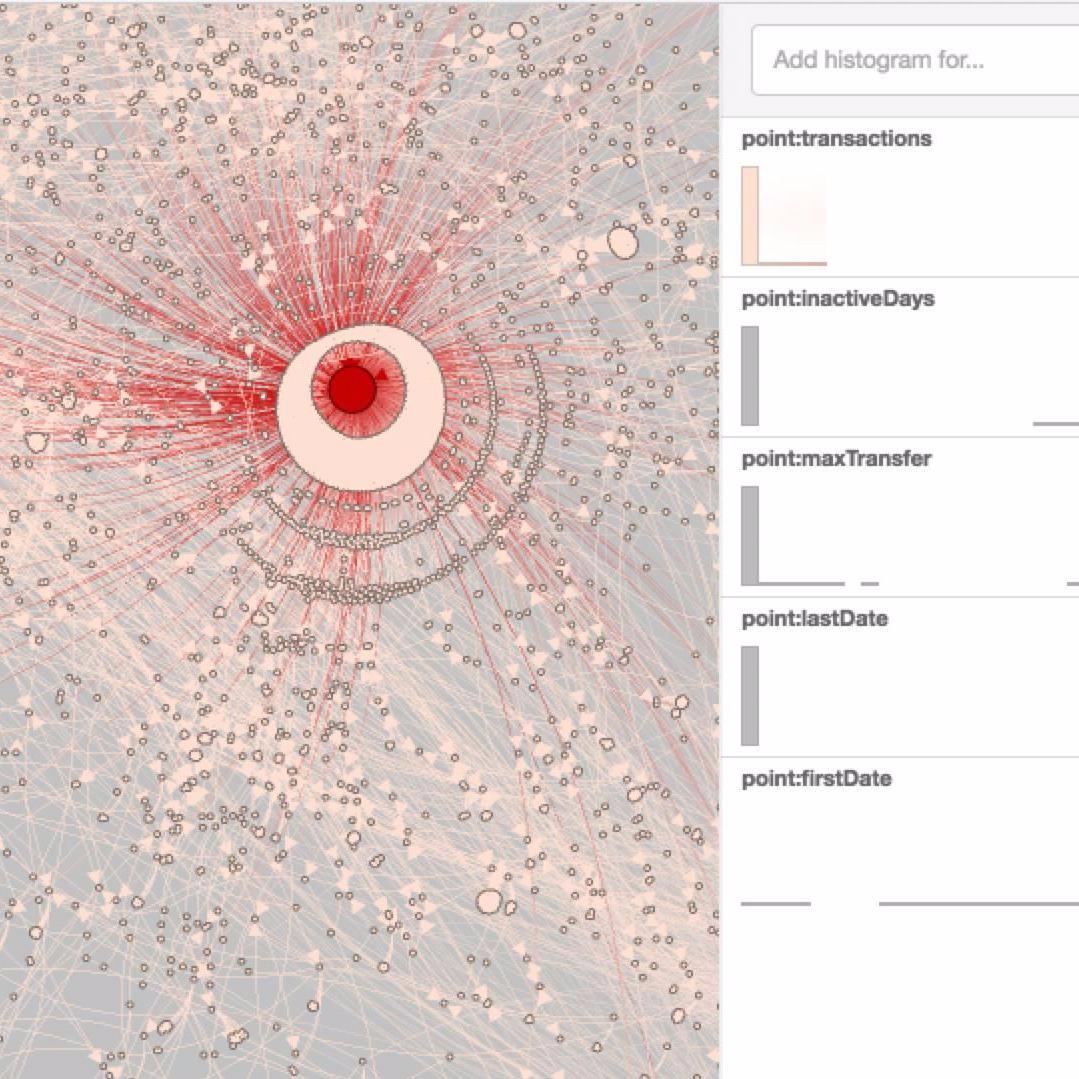 |
Port Scan Attack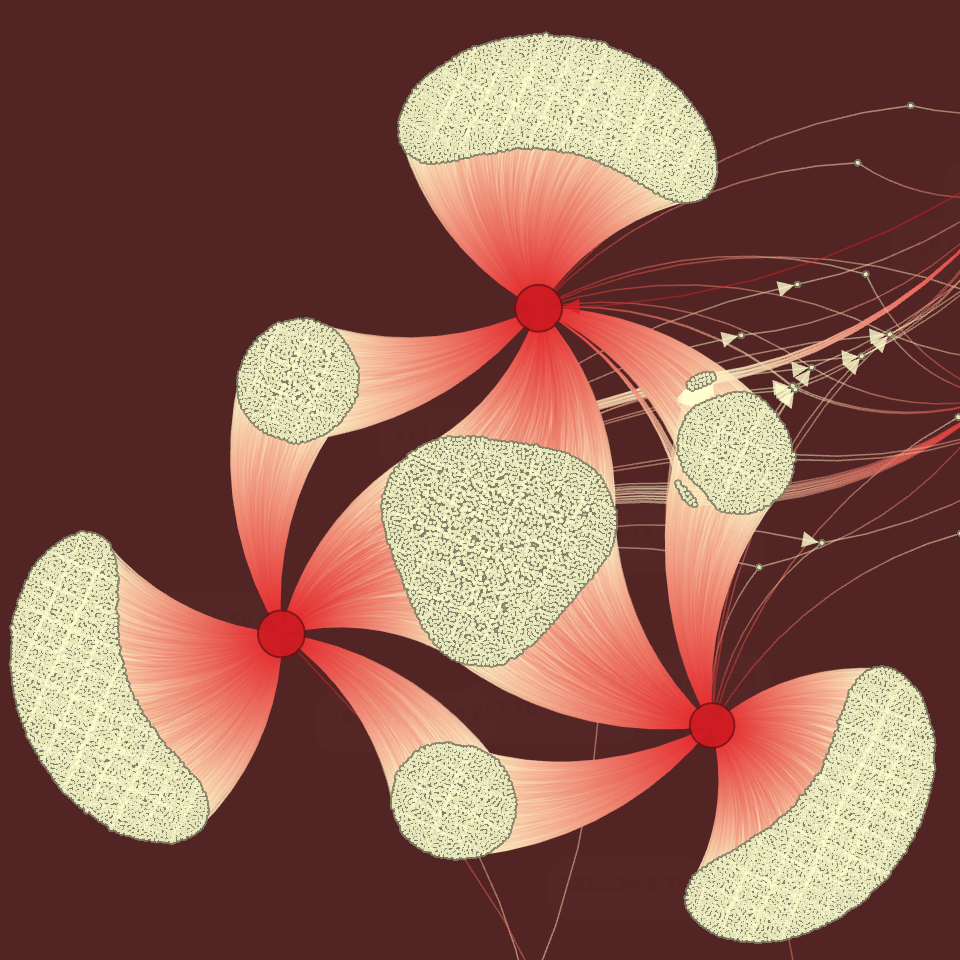 |
Protein Interactions 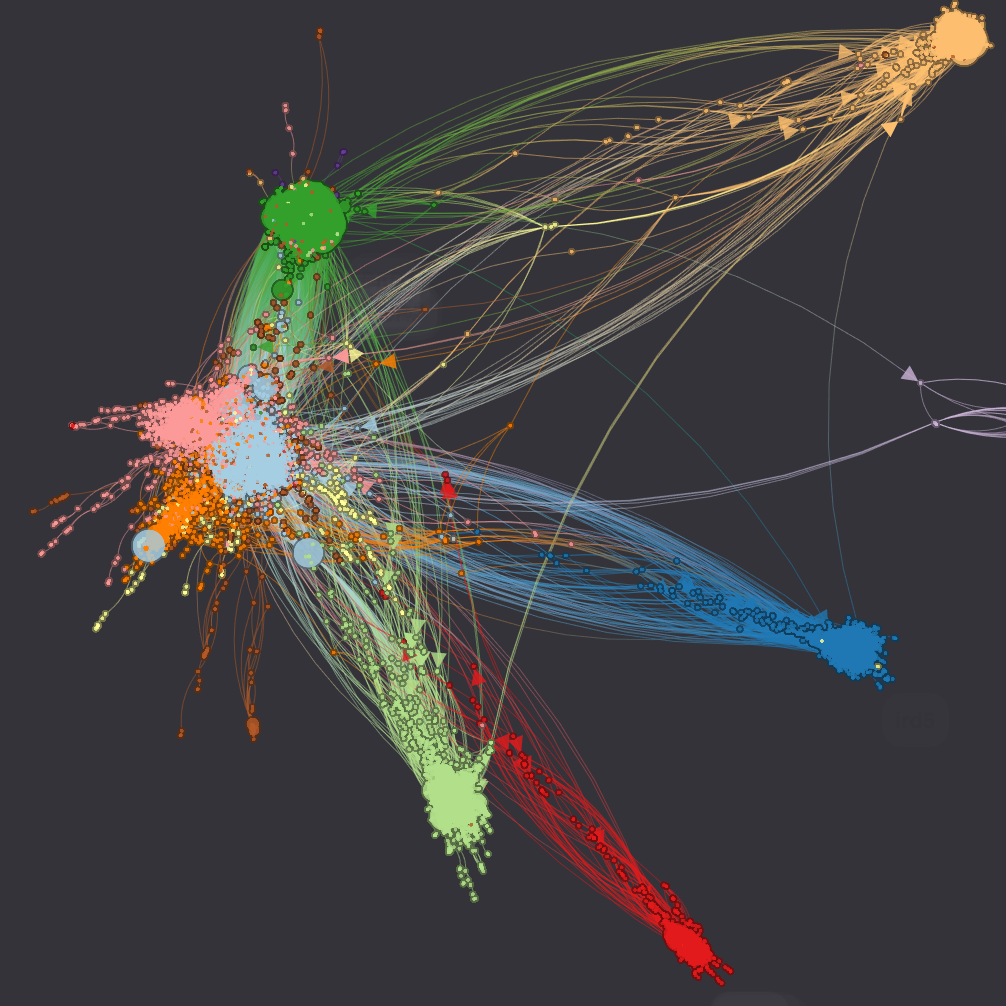 (data) (data) |
Programming Languages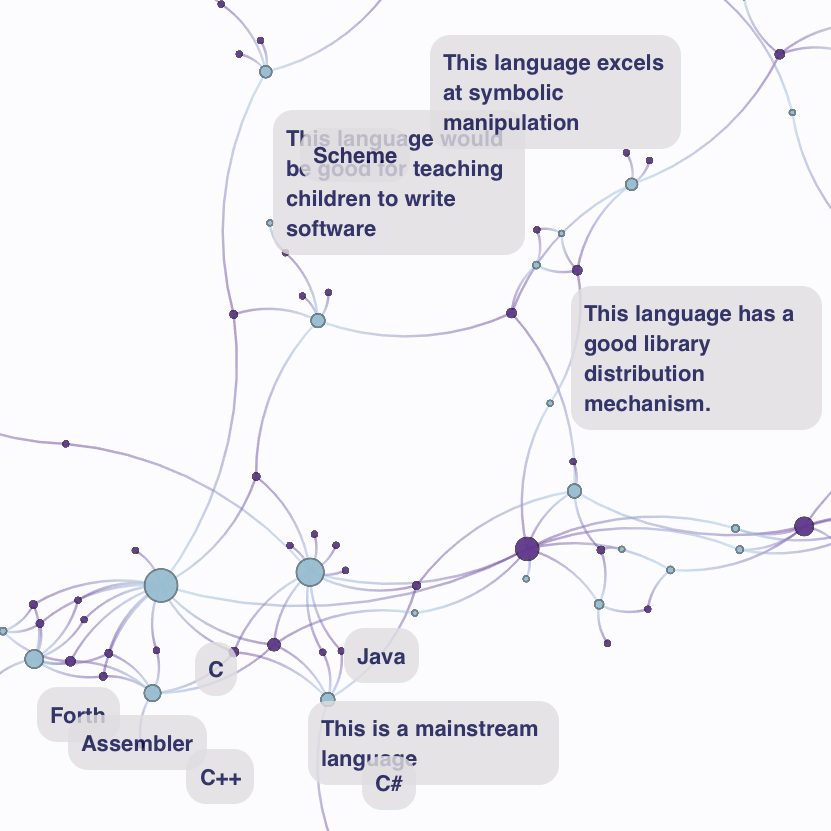 (data) (data) |
Install
Common configurations:
-
Minimal core
Includes: The GFQL dataframe-native graph query language, built-in layouts, Graphistry visualization server client
pip install graphistry
Does not include
graphistry[ai], plugins -
No dependencies and user-level
pip install --no-deps --user graphistry
-
GPU acceleration - Optional
Local GPU: Install RAPIDS and/or deploy a GPU-ready Graphistry server
Remote GPU: Use the remote endpoints.
For further options, see the installation guides
Visualization quickstart
Quickly go from raw data to a styled and interactive Graphistry graph visualization:
import graphistry
import pandas as pd
# Raw data as Pandas CPU dataframes, cuDF GPU dataframes, Spark, ...
df = pd.DataFrame({
'src': ['Alice', 'Bob', 'Carol'],
'dst': ['Bob', 'Carol', 'Alice'],
'friendship': [0.3, 0.95, 0.8]
})
# Bind
g1 = graphistry.edges(df, 'src', 'dst')
# Override styling defaults
g1_styled = g1.encode_edge_color('friendship', ['blue', 'red'], as_continuous=True)
# Connect: Free GPU accounts and self-hosting @ graphistry.com/get-started
graphistry.register(api=3, username='your_username', password='your_password')
# Upload for GPU server visualization session
g1_styled.plot()
Explore 10 Minutes to Graphistry Visualization for more visualization examples and options
PyGraphistry[AI] & GFQL quickstart - CPU & GPU
CPU graph pipeline combining graph ML, AI, mining, and visualization:
from graphistry import n, e, e_forward, e_reverse
# Graph analytics
g2 = g1.compute_igraph('pagerank')
assert 'pagerank' in g2._nodes.columns
# Graph ML/AI
g3 = g2.umap()
assert ('x' in g3._nodes.columns) and ('y' in g3._nodes.columns)
# Graph querying with GFQL
g4 = g3.chain([
n(query='pagerank > 0.1'), e_forward(), n(query='pagerank > 0.1')
])
assert (g4._nodes.pagerank > 0.1).all()
# Upload for GPU server visualization session
g4.plot()
The automatic GPU modes require almost no code changes:
import cudf
from graphistry import n, e, e_forward, e_reverse
# Modified -- Rebind data as a GPU dataframe and swap in a GPU plugin call
g1_gpu = g1.edges(cudf.from_pandas(df))
g2 = g1_gpu.compute_cugraph('pagerank')
# Unmodified -- Automatic GPU mode for all ML, AI, GFQL queries, & visualization APIs
g3 = g2.umap()
g4 = g3.chain([
n(query='pagerank > 0.1'), e_forward(), n(query='pagerank > 0.1')
])
g4.plot()
Explore 10 Minutes to PyGraphistry for a wider variety of graph processing.
PyGraphistry documentation
- Main PyGraphistry documentation
- 10 Minutes to: PyGraphistry, Visualization, GFQL
- Get started: Install, UI Guide, Notebooks
- Performance: PyGraphistry CPU+GPU & GFQL CPU+GPU
- API References
- PyGraphistry API Reference: Visualization & Compute, PyGraphistry Cheatsheet
- GFQL Documentation: GFQL Cheatsheet and GFQL Operator Cheatsheet
- Plugins: Databricks, Splunk, Neptune, Neo4j, RAPIDS, and more
- Web: iframe, JavaScript, REST
Graphistry ecosystem
-
Graphistry server:
- Launch - Graphistry Hub, Graphistry cloud marketplaces, and self-hosting
- Self-hosting: Administration (including Docker) & Kubernetes
-
Graphistry client APIs:
-
Additional projects:
- Louie.ai: GenAI-native notebooks & dashboards to talk to your databases & Graphistry
- graph-app-kit: Streamlit Python dashboards with batteries-include graph packages
- cu-cat: Automatic GPU feature engineering
Community and support
- Blog for tutorials, case studies, and updates
- Slack: Join the Graphistry Community Slack for discussions and support
- Twitter & LinkedIn: Follow for updates
- GitHub Issues open source support
- Graphistry ZenDesk dedicated enterprise support
Contribute
See CONTRIBUTING and DEVELOP for participating in PyGraphistry development, or reach out to our team
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file graphistry-0.50.6.tar.gz.
File metadata
- Download URL: graphistry-0.50.6.tar.gz
- Upload date:
- Size: 404.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
1ec6fc2ff5d3d3943257f508f15a3de866da31de657895dcf95624caaefaa357
|
|
| MD5 |
01291b8398d73c505c6f7eadcdd43a49
|
|
| BLAKE2b-256 |
478b1cbe6984b8ed78e709aa9616a981d21ab68c817b2b19b61536470166b3a0
|
File details
Details for the file graphistry-0.50.6-py3-none-any.whl.
File metadata
- Download URL: graphistry-0.50.6-py3-none-any.whl
- Upload date:
- Size: 439.8 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
5a69d746e9c8a8f37f151b0aed7f80351ad07c3724fe25f678b201b2bab728e7
|
|
| MD5 |
c2c59d0cd497084d49bad5a29b8526db
|
|
| BLAKE2b-256 |
8724b8534027c5a49fcc73f69172b0715fc053303b496c122a874d51e6ebd006
|





































