A data integration algorithm.
Project description
harmonypy
harmonypy is a Python implementation of the Harmony algorithm for integrating multiple high-dimensional datasets.
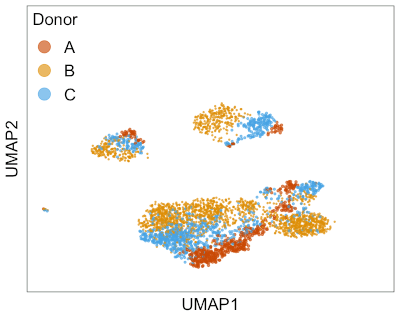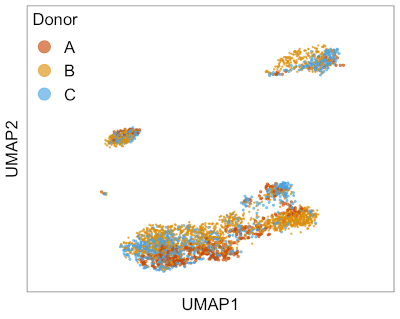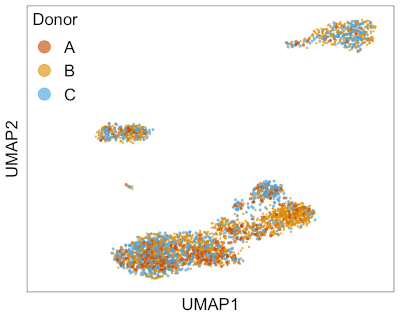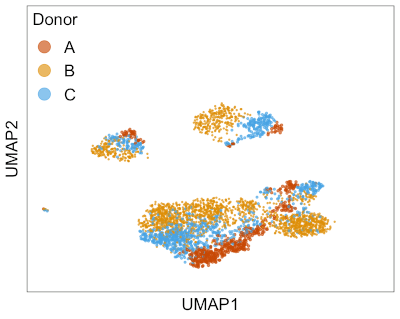
This animation shows Harmony aligning three single-cell RNA-seq datasets from different donors. → How to make this animation. Before Harmony, you can clearly distinguish cells from each of the three donors. After Harmony, the cells from different donors are mixed while preserving the overall shape of the data. This makes it easier to run clustering algorithms to find similar cell types that are present in different batches of data.
Installation
pip install harmonypy
Quick Start
import harmonypy as hm
import pandas as pd
# Load the principal components and metadata
pcs = pd.read_csv("data/pbmc_3500_pcs.tsv.gz", sep="\t")
meta = pd.read_csv("data/pbmc_3500_meta.tsv.gz", sep="\t")
# Run Harmony to correct for batch effects (donor)
harmony_out = hm.run_harmony(pcs, meta, "donor")
# Save corrected PCs (same shape as input)
result = pd.DataFrame(harmony_out.Z_corr, columns=pcs.columns)
result.to_csv("pbmc_3500_pcs_harmony.tsv", sep="\t", index=False)
Performance
Apple M1 Ultra (2022) with PyTorch MPS backend:
Small (3.5k cells x 30 PCs): 3.48s
Medium (69k cells x 50 PCs): 9.26s
Large (858k cells x 29 PCs): 21.75s
Note: For small datasets, the NumPy-only version (v0.1.0) may be faster due to GPU overhead.
Citation
If you use Harmony in your work, please cite the original paper:
Korsunsky, I., Millard, N., Fan, J. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296 (2019). https://doi.org/10.1038/s41592-019-0619-0
The Supplementary Information PDF provides detailed mathematical descriptions and implementation notes.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file harmonypy-0.2.0.tar.gz.
File metadata
- Download URL: harmonypy-0.2.0.tar.gz
- Upload date:
- Size: 22.5 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: uv/0.9.10 {"installer":{"name":"uv","version":"0.9.10"},"python":null,"implementation":{"name":null,"version":null},"distro":{"name":"macOS","version":null,"id":null,"libc":null},"system":{"name":null,"release":null},"cpu":null,"openssl_version":null,"setuptools_version":null,"rustc_version":null,"ci":null}
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
9678a059f943723ba954f58d04e1a72e8a5c49ef4d95ba04fb33c9c1b5c67a77
|
|
| MD5 |
bb11f230802938e1be5094c9d9059cf2
|
|
| BLAKE2b-256 |
7374976a4a6cc4933f578228ea330052c700a4057f7d04540ae05d461bee4aaa
|
File details
Details for the file harmonypy-0.2.0-py3-none-any.whl.
File metadata
- Download URL: harmonypy-0.2.0-py3-none-any.whl
- Upload date:
- Size: 23.1 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: uv/0.9.10 {"installer":{"name":"uv","version":"0.9.10"},"python":null,"implementation":{"name":null,"version":null},"distro":{"name":"macOS","version":null,"id":null,"libc":null},"system":{"name":null,"release":null},"cpu":null,"openssl_version":null,"setuptools_version":null,"rustc_version":null,"ci":null}
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
663b679701f974fff03206f1188066e4fc0f38d432ea43cf0ee143f65c95cf8b
|
|
| MD5 |
c71ccfd8bb363da2422c934ad185824c
|
|
| BLAKE2b-256 |
0fb2e9a468d388b772e35f0918a20bd03812a5d83fb776c8b509e79209d87c05
|















