File organization made easy using tags
Project description
HyperTag
File organization made easy. HyperTag let's humans intuitively express how they think about their files using tags.
Objective Function: Minimize the time between a thought and access to all your relevant files.
Install
Available on PyPI
$ pip install hypertag
Overview
HyperTag offers a slick CLI but more importantly it creates a directory called HyperTagFS which is a file system based representation of your files and tags using symbolic links and directories.
Directory Import: Import your existing directory hierarchies using $ hypertag import path/to/directory. HyperTag converts it automatically into a tag hierarchy using metatagging.
Semantic Search (Experimental): Search through all your text documents (yes, even PDF's) content. This function is powered by the awesome Sentence Transformers library. Currently only English is supported.
HyperTag Daemon (Experimental): Monitors HyperTagFS for user changes. Currently supports file and directory (tag) deletions + directory (name as query) creation with automatic query result population. Also spawns the DaemonService which speeds up semantic search significantly.
Fuzzy Matching Queries: HyperTag uses fuzzy matching to minimize friction in the unlikely case of a typo.
File Type Groups: HyperTag automatically creates folders containing common files (e.g. Images: jpg, png, etc., Documents: txt, pdf, etc., Source Code: py, js, etc.), which can be found in HyperTagFS.
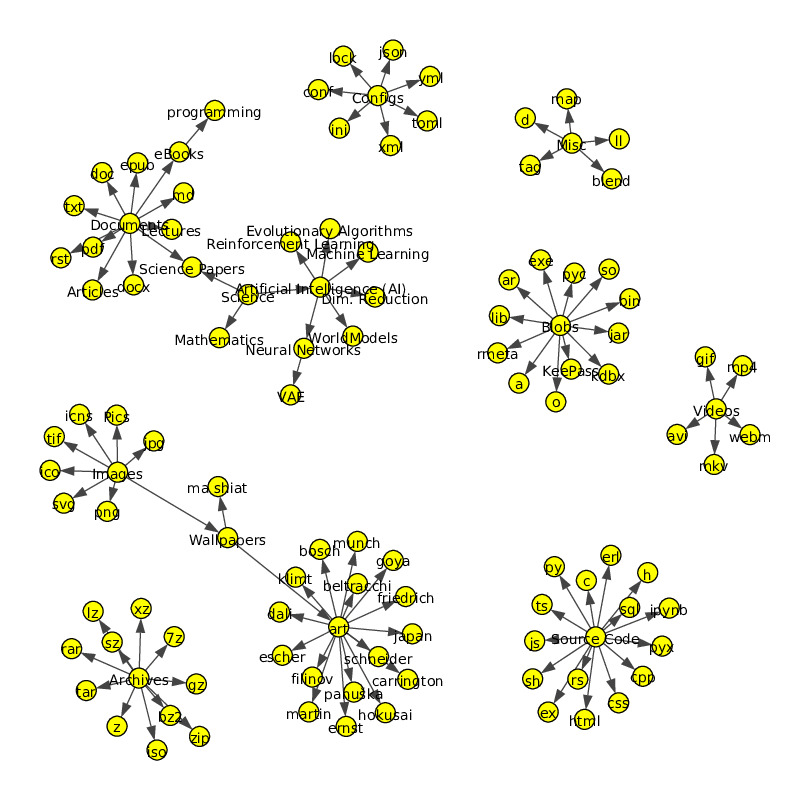
HyperTag Graph: Quickly get an overview of your HyperTag Graph! HyperTag visualizes the metatag graph on every change and saves it at HyperTagFS/hypertag-graph.pdf.
CLI Functions
Import existing directory recursively
Import files with tags inferred from the existing directory hierarchy
$ hypertag import path/to/directory
Tag file/s
Manually tag files
$ hypertag tag humans/*.txt with human "Homo Sapiens"
Untag file/s
Manually remove tag/s from file/s
$ hypertag untag humans/*.txt with human "Homo Sapiens"
Tag a tag
Metatag tag/s to create tag hierarchies
$ hypertag metatag human with animal
Merge tags
Merge all associations (files & tags) of tag A into tag B
$ hypertag merge human into "Homo Sapiens"
Query using Set Theory
Print file names of the resulting set matching the query. Queries are composed of tags and operands. Tags are fuzzy matched for convenience. Nesting is currently not supported, queries are evaluated from left to right
Print paths: $ hypertag query human --path
Print fuzzy matched tag: $ hypertag query man --verbose
Disable fuzzy matching: $ hypertag query human --fuzzy=0
Default operand is AND (intersection):
$ hypertag query human "Homo Sapiens" is equivalent to $ hypertag query human and "Homo Sapiens"
OR (union):
$ hypertag query human or "Homo Sapiens"
MINUS (difference):
$ hypertag query human minus "Homo Sapiens"
Index available text files
Only indexed files can be searched.
To parse even unparseable PDF's, install tesseract: # pacman -S tesseract tesseract-data-eng
$ hypertag index
Semantic search indexed text files
Print file names sorted by matching score. Performance benefits greatly from running the HyperTag daemon.
$ hypertag search "your important text query"
Print all tags of file/s
$ hypertag tags filename1 filename2
Print all metatags of tag/s
$ hypertag metatags tag1 tag2
Print all tags
$ hypertag show
Print all files
Print names:
$ hypertag show files
Print paths:
$ hypertag show files --path
Visualize HyperTag Graph
Visualize the metatag graph hierarchy (saved at HyperTagFS root)
$ hypertag graph
Specify layout algorithm (default: fruchterman_reingold):
$ hypertag graph --layout=kamada_kawai
Generate HyperTagFS
Generate file system based representation of your files and tags using symbolic links and directories
$ hypertag mount
Start HyperTag daemon
Start daemon process with dual function:
- Watch HyperTagFS directory for user changes
- Spawn DaemonService to load and expose model used for semantic search
$ hypertag daemon
Set HyperTagFS directory path
Default is the user's home directory
$ hypertag set_hypertagfs_dir path/to/directory
Architecture
- Python powers HyperTag
- Many other awesome open-source projects make HyperTag possible (listed in
pyproject.toml) - SQLite3 serves as the meta data storage engine (located at
~/.config/hypertag/hypertag.db) - Symbolic links are used to create the HyperTagFS directory structure
- Semantic text document search is powered by the awesome DistilBERT
Development
- Clone repo:
$ git clone https://github.com/SeanPedersen/HyperTag.git $ cd HyperTag/- Install Poetry
- Install dependencies:
$ poetry install - Activate virtual environment:
$ poetry shell - Run all tests:
$ pytest -v - Run formatter:
$ black hypertag/ - Run linter:
$ flake8 - Run type checking:
$ mypy **/*.py - Run security checking:
$ bandit --exclude tests/ -r . - Run HyperTag:
$ python -m hypertag
Inspiration
What is the point of HyperTag's existence?
HyperTag offers many unique features such as the import, semantic search, graphing and fuzzy matching functions that make it very convenient to use. All while HyperTag's code base staying tiny at <1000 LOC in comparison to TMSU (>10,000 LOC) and SuperTag (>25,000 LOC), making it easy to hack on.
This project is partially inspired by these open-source projects:
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for hypertag-0.4.3.1-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 7f67841c30d195e13af5fcf34b155fb1e1152d8691c3325a5db4e34e056a9180 |
|
| MD5 | bfa65cd7800aa226dd586d71f7e1f7f6 |
|
| BLAKE2b-256 | aab968cf13e413a513c237dfd86c7f1996ef5607bef45a98b86410d2bf215d34 |