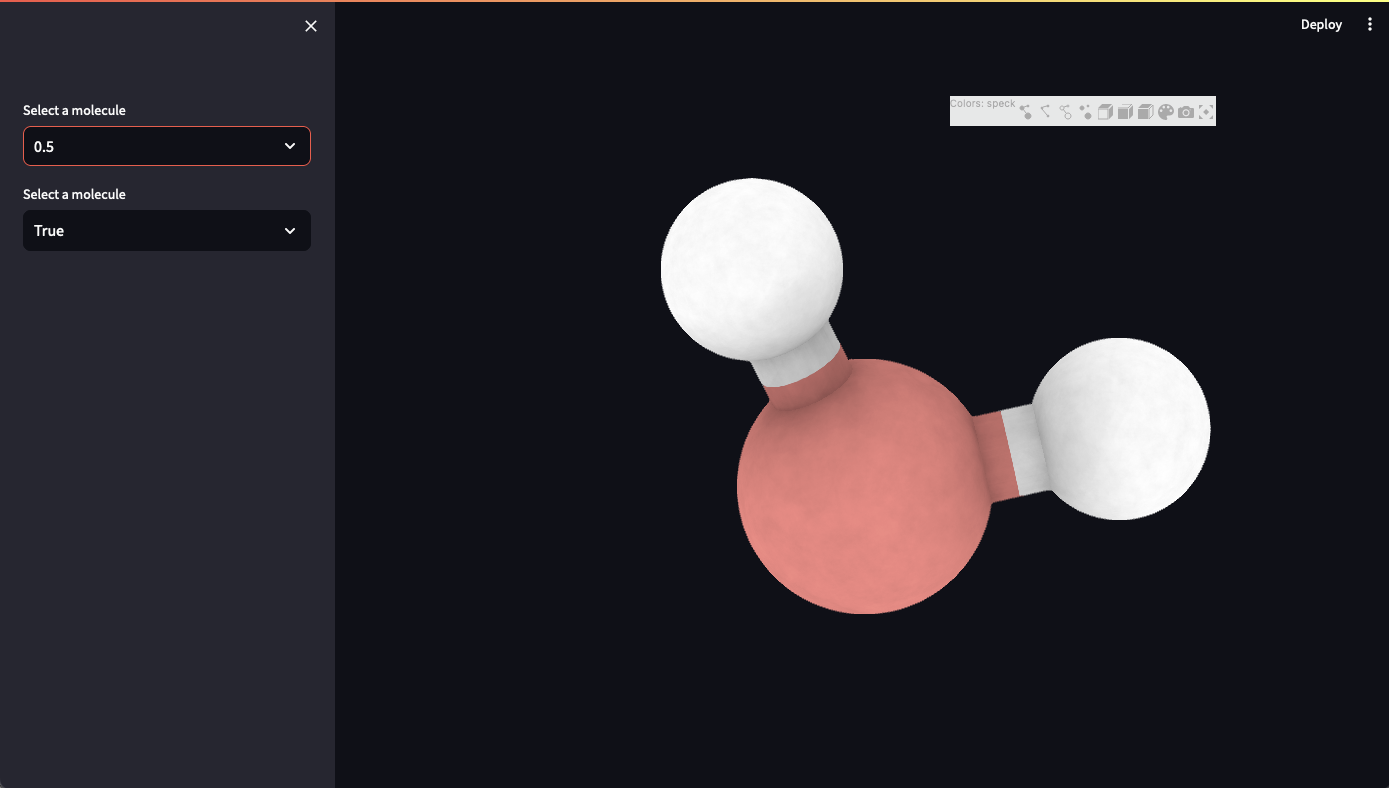
Speck Jupyter Widget
Project description
ipyspeck
A Jupyter Widget for rendering beautiful molecular structures using Speck.
| Latest Release |

|
| PyPI Downloads |


|
About
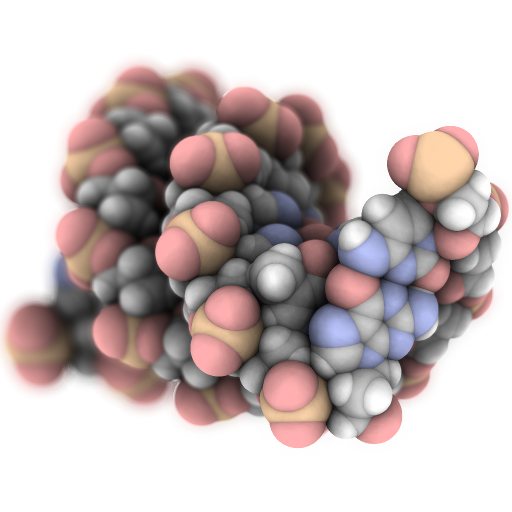
Speck is a molecule renderer with the goal of producing figures that are as attractive as they are practical. Express your molecule clearly and with style.
Ipyspeck is an ipywidget wrapper for Speck that allows you to visualize molecular structures directly in Jupyter notebooks and JupyterLab.
Version Compatibility
⚠️ IMPORTANT: Version Compatibility Notice
ipyspeck 0.7.x and later requires:
- JupyterLab >= 3.0
- ipywidgets >= 7.0
- Python >= 3.9
For older environments, use ipyspeck 0.6.x:
- JupyterLab 2.x → use
ipyspeck<0.7- ipywidgets < 7.0 → use
ipyspeck<0.7- Python < 3.9 → use
ipyspeck<0.7Migration Notes:
- Version 0.7.0+ uses the modern JupyterLab 3+ federated extension system
- Version 0.7.0+ supports both ipywidgets 7.x and 8.x with backward compatibility
- Version 0.7.0+ uses Lumino (LuminoJS) instead of deprecated PhosphorJS
Installation
Standard Installation
For JupyterLab 3+ and ipywidgets 7+:
pip install ipyspeck
That's it! The extension will be automatically enabled in JupyterLab 3+.
Legacy Installation (JupyterLab 2.x)
For older JupyterLab versions:
pip install "ipyspeck<0.7"
jupyter nbextension enable --py --sys-prefix ipyspeck
jupyter labextension install ipyspeck
Development Installation
For developers who want to contribute:
git clone https://github.com/denphi/speck.git
cd speck/widget/ipyspeck
pip install -e .
Usage
Basic Usage
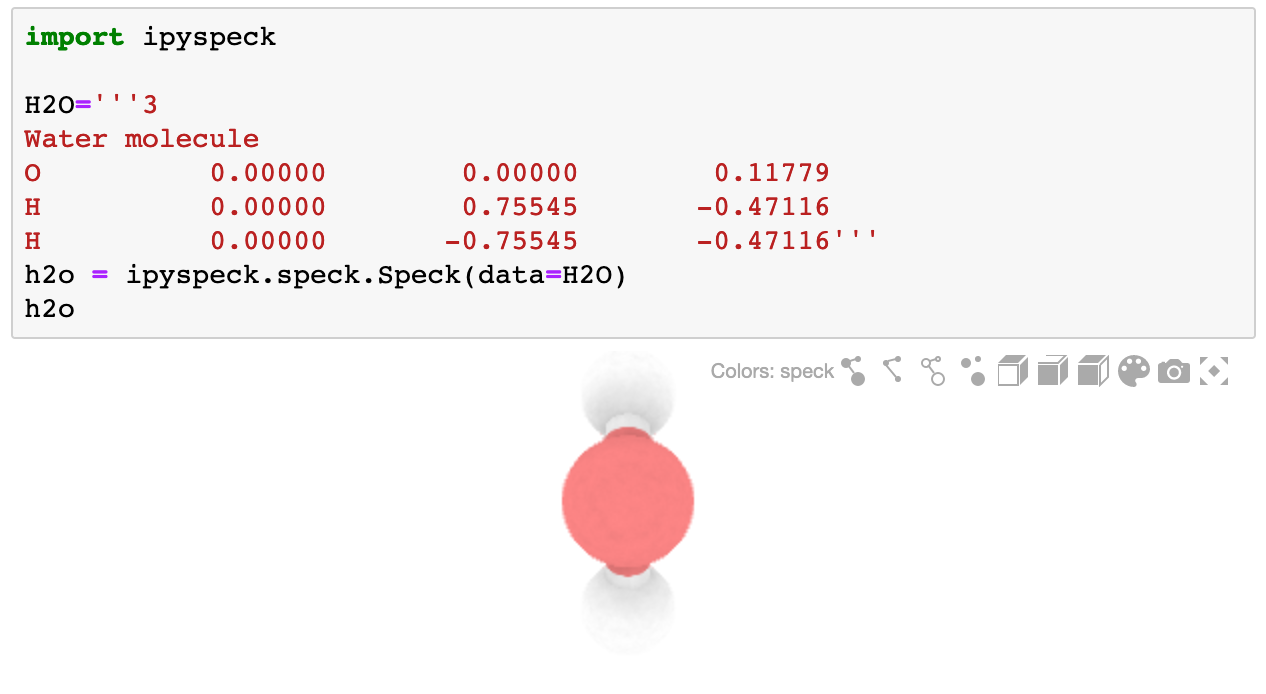
The ipyspeck widget renders molecules in XYZ format:
from ipyspeck import Speck
H2O = '''3
Water molecule
O 0.00000 0.00000 0.11779
H 0.00000 0.75545 -0.47116
H 0.00000 -0.75545 -0.47116'''
h2o = Speck(data=H2O)
h2o
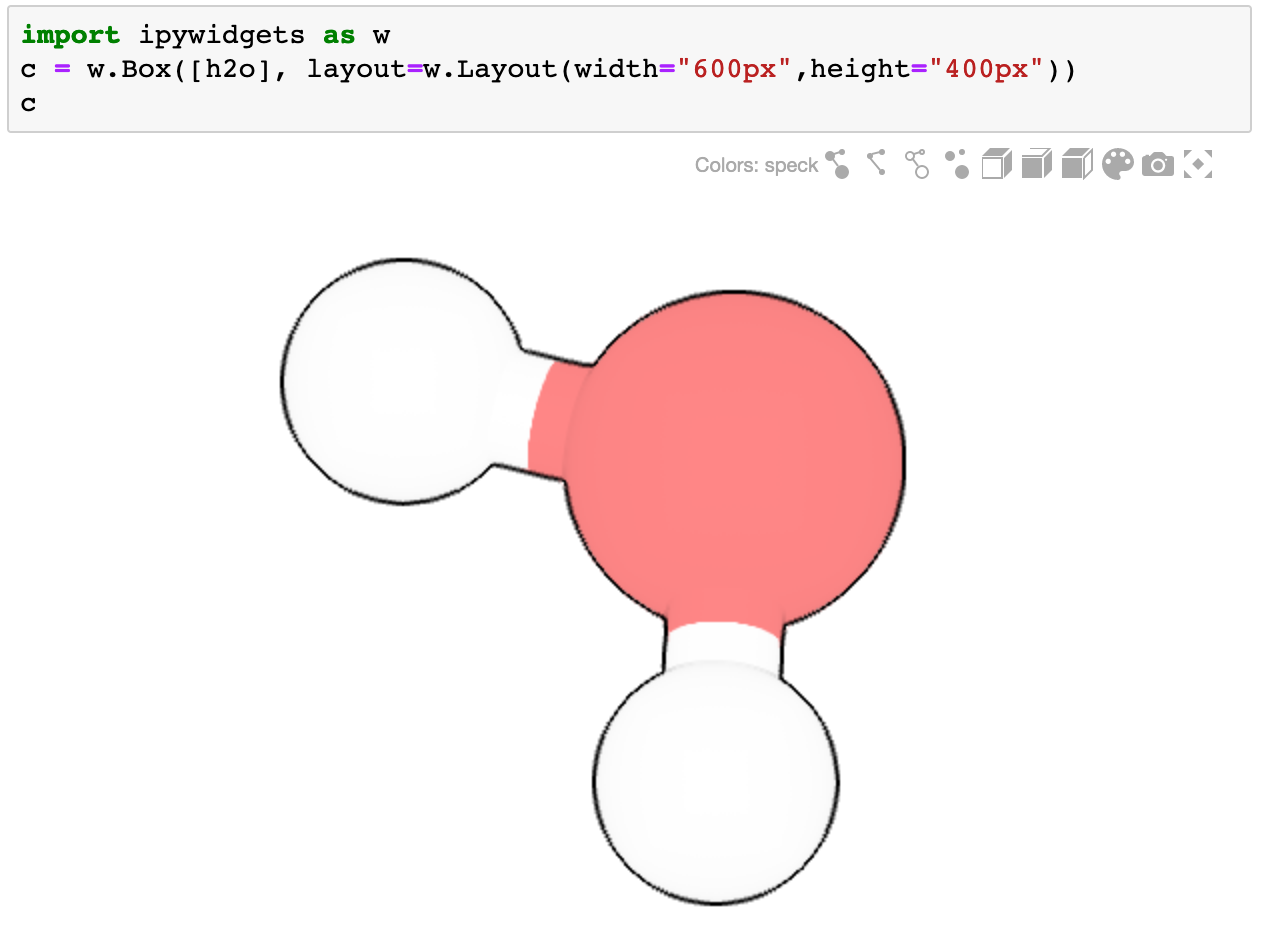
Using with Container Widgets
For better control over size and layout, use ipyspeck inside container widgets:
import ipywidgets as widgets
# Create a sized container
container = widgets.Box(
[h2o],
layout=widgets.Layout(width="600px", height="400px")
)
container
Customization
Adjust visualization parameters to suit your needs:
# Modify atom size
h2o.atomScale = 0.3
# Change bond thickness
h2o.bondScale = 0.3
# Toggle atom outlines
h2o.outline = 0
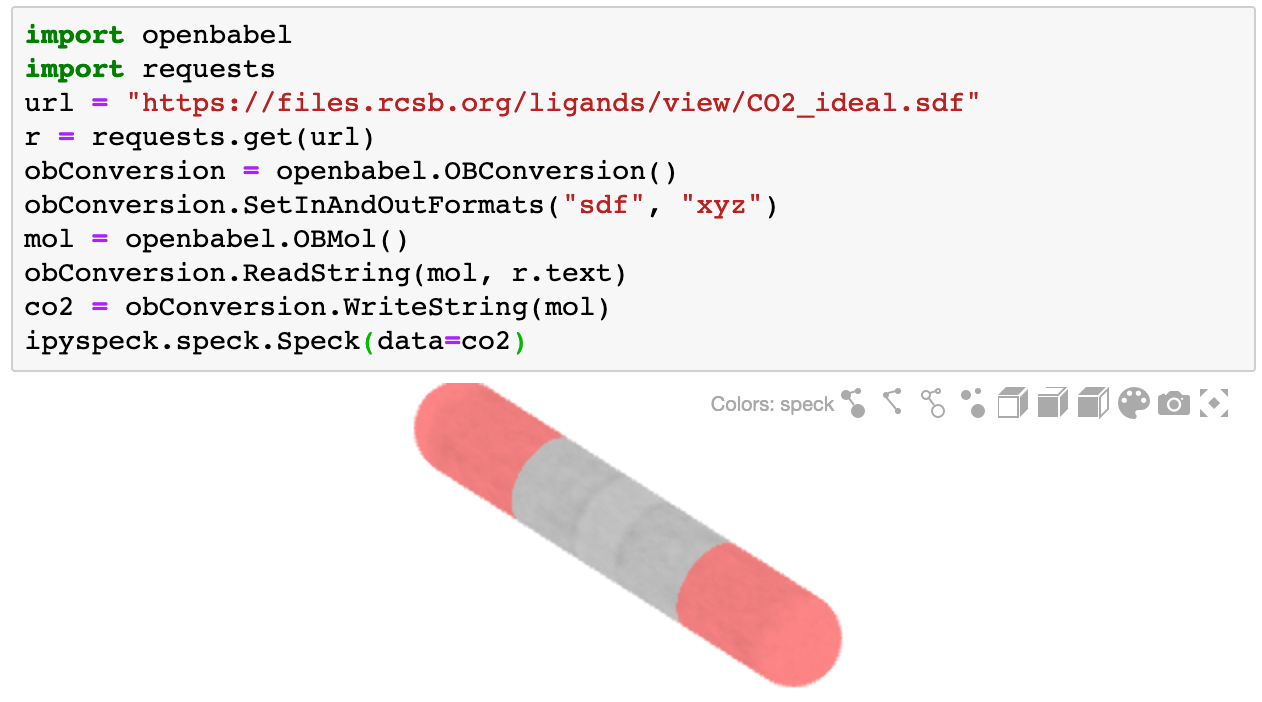
Working with Different File Formats
Use OpenBabel to convert various molecular formats to XYZ:
import openbabel
import requests
# Fetch a molecule in SDF format
url = "https://files.rcsb.org/ligands/view/CO2_ideal.sdf"
r = requests.get(url)
# Convert to XYZ
obConversion = openbabel.OBConversion()
obConversion.SetInAndOutFormats("sdf", "xyz")
mol = openbabel.OBMol()
obConversion.ReadString(mol, r.text)
co2 = obConversion.WriteString(mol)
# Visualize
Speck(data=co2)
Streamlit Integration
ipyspeck 0.6+ includes a Streamlit wrapper for building interactive web apps:
import streamlit as st
from ipyspeck import stspeck
H2O = '''3
Water molecule
O 0.00000 0.00000 0.11779
H 0.00000 0.75545 -0.47116
H 0.00000 -0.75545 -0.47116'''
with st.sidebar:
ao = st.selectbox("Ambient Occlusion", [0, 0.1, 0.2, 0.5, 0.8, 1])
bonds = st.selectbox("Show Bonds", [True, False])
res = stspeck.Speck(
data=H2O,
ao=ao,
bonds=bonds,
width="800px",
height="600px"
)
Features
- 🎨 Beautiful, publication-quality molecular visualizations
- 🔄 Interactive rotation and zoom
- ⚡ Fast rendering with WebGL
- 🎛️ Customizable atom and bond styles
- 📦 Support for XYZ format (use OpenBabel for other formats)
- 🧪 Works in Jupyter Notebook, JupyterLab, and Streamlit
- 🔌 Compatible with ipywidgets ecosystem
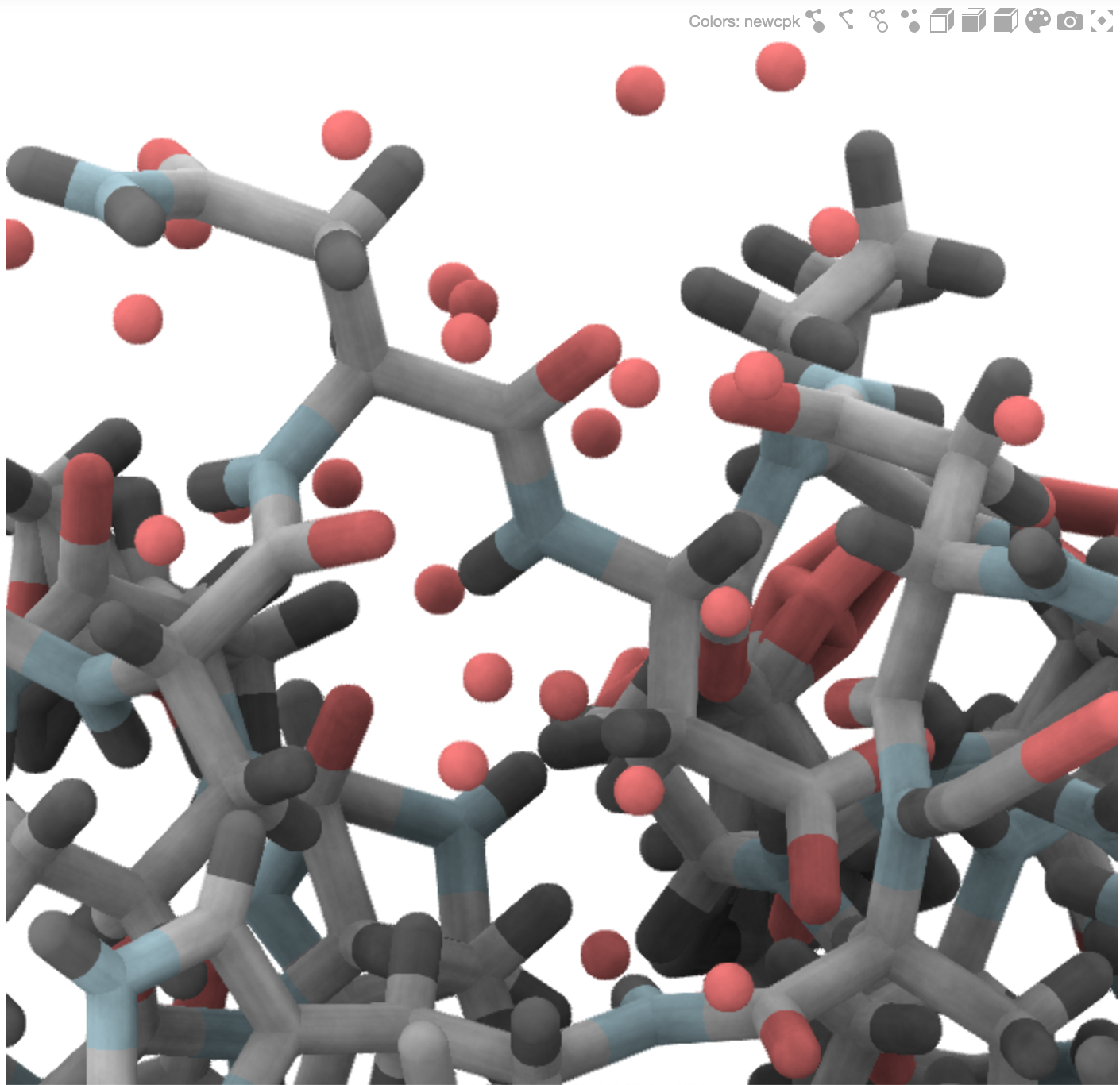
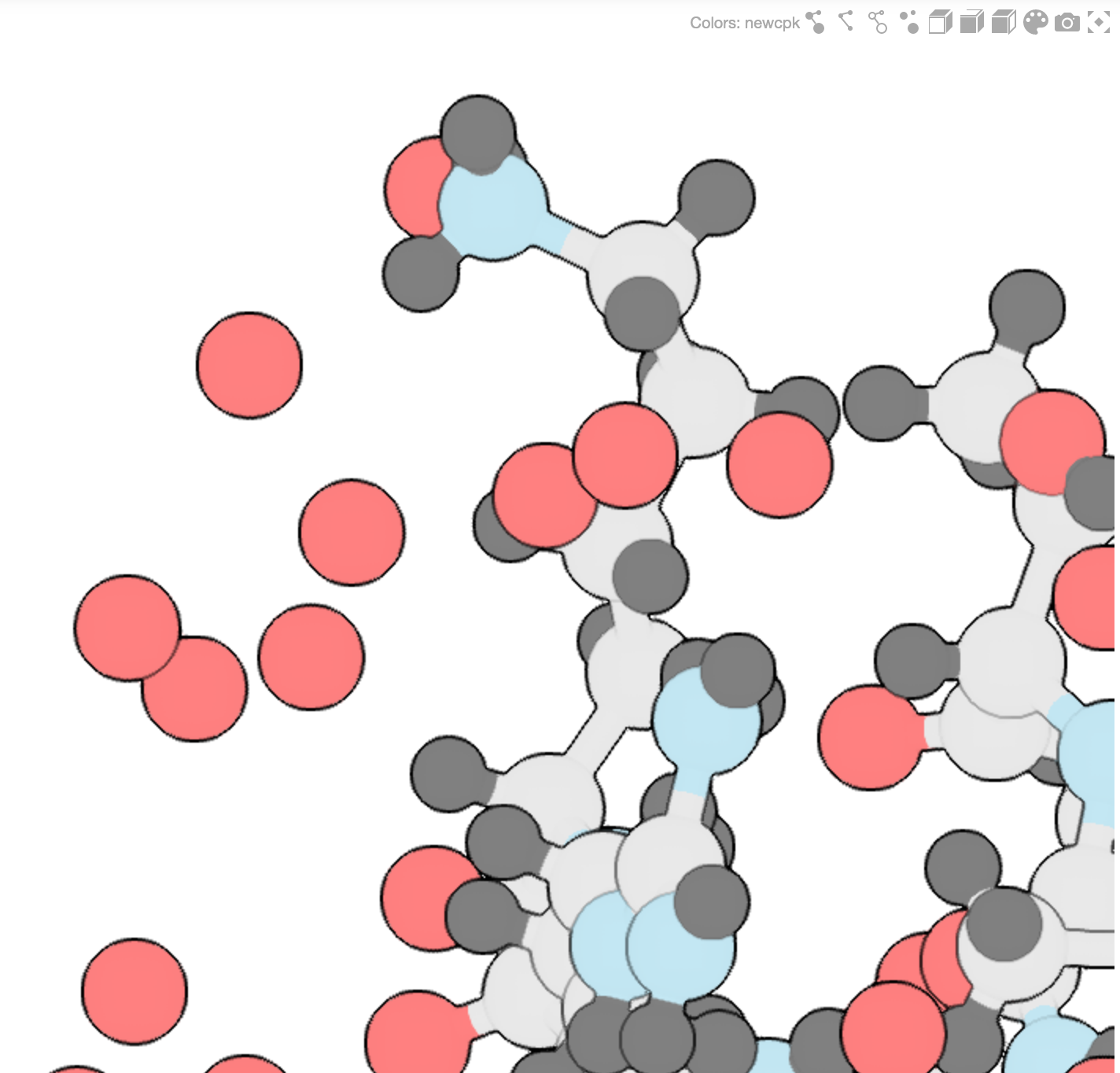
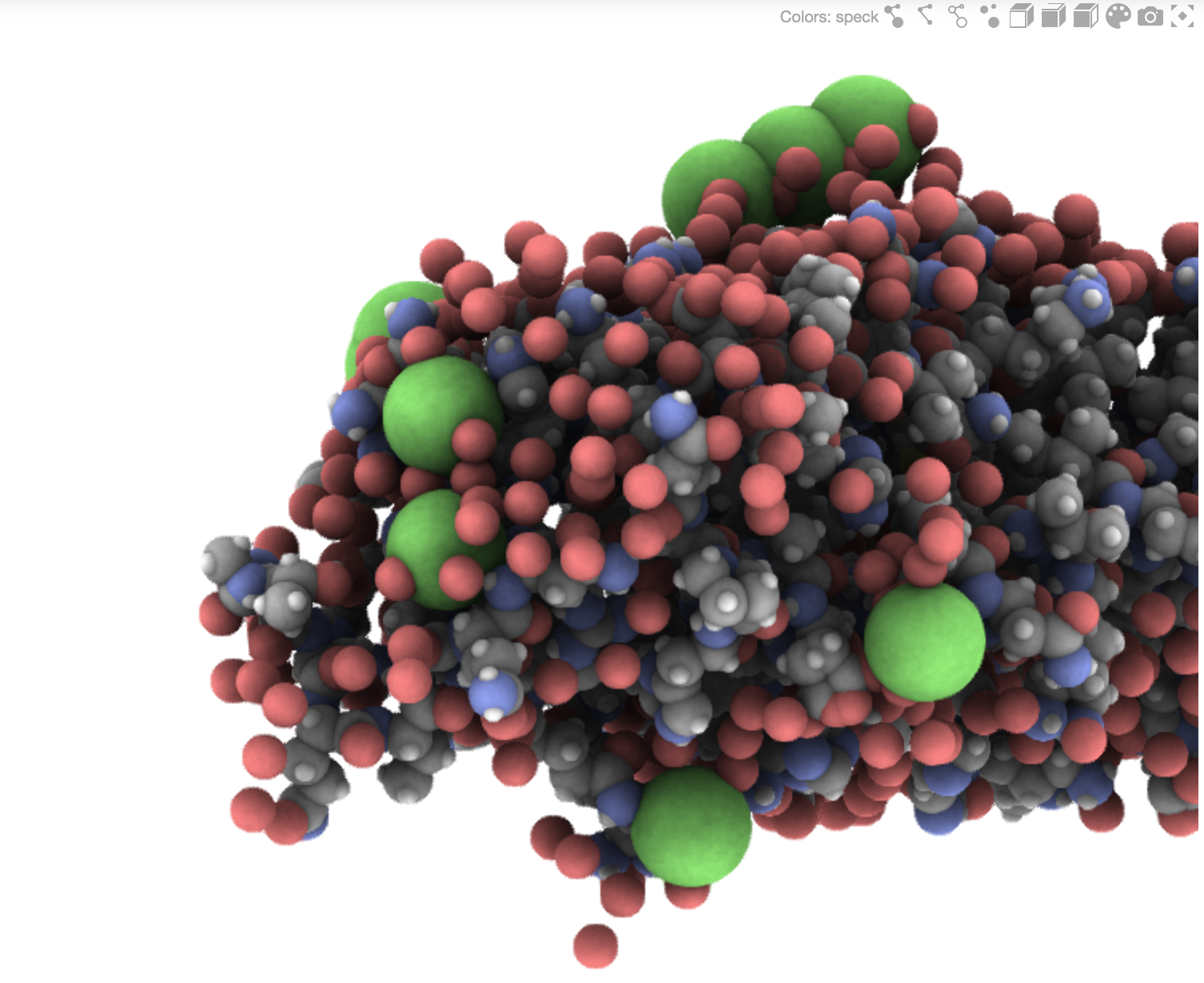
Gallery


Development
Building from Source
# Install dependencies
npm install
# Build TypeScript and JavaScript
npm run build
# Or build for production
npm run build:prod
Active Development
When actively developing the extension:
# Watch for changes and rebuild automatically
jupyter lab --watch
Note: On first jupyter lab --watch, you may need to touch a file to trigger JupyterLab to open.
License
BSD-3-Clause
Author
Daniel Mejia (Denphi) - denphi@denphi.com
Links
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file ipyspeck-0.7.1.tar.gz.
File metadata
- Download URL: ipyspeck-0.7.1.tar.gz
- Upload date:
- Size: 23.7 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.10.11
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
26c7f191b64d3ae46db6485a093ddb402ec7c0258706193b1909b0492dce5f5a
|
|
| MD5 |
ffe2bdb5e169531c06f2913658aa29f5
|
|
| BLAKE2b-256 |
2c1a21b2c83ac62e136d7a01d41d212a59a9d558e96f496dccbc24c290e45aaa
|
File details
Details for the file ipyspeck-0.7.1-py3-none-any.whl.
File metadata
- Download URL: ipyspeck-0.7.1-py3-none-any.whl
- Upload date:
- Size: 939.9 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.10.11
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
73fd14d0e974a6931bb6665205b59c8ef5c3019022470ab84ebcc199a259956e
|
|
| MD5 |
10da8a4ae1d093022481d957a2e80e0b
|
|
| BLAKE2b-256 |
71c1cf643efbdee3595ccc2f86c7201dc74fb271064080d430c4c9ae8fac6c9c
|