Keras models with TQDM progress bars in Jupyter notebooks
Project description
Keras integration with TQDM progress bars.
Keras is an awesome machine learning library for Theano or TensorFlow.
TQDM is a progress bar library with good support for nested loops and Jupyter/IPython notebooks.
Key features
TQDM supports nested progress bars. If you have Keras fit and predict loops within an outer TQDM loop, the nested loops will display properly.
TQDM supports Jupyter/IPython notebooks.
TQDM looks great!
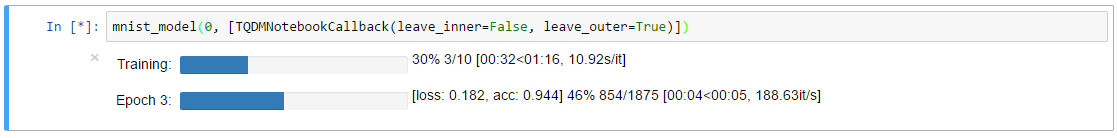
TQDMNotebookCallback with leave_inner=False (default)
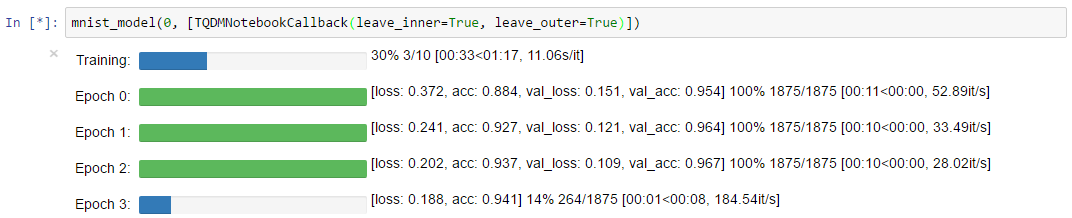
TQDMNotebookCallback with leave_inner=True
TQDMCallback for command-line scripts

Installation
Stable release
pip install keras-tqdm
Development release
pip install git+https://github.com/bstriner/keras-tqdm.git --upgrade --no-deps
Development mode (changes to source take effect without reinstalling)
git clone https://github.com/bstriner/keras-tqdm.git cd keras-tqdm python setup.py develop
Basic usage
It’s very easy to use Keras TQDM. The only required change is to remove default messages (verbose=0) and add a callback to model.fit. The rest happens automatically! For Jupyter Notebook required code modification is as simple as:
from keras_tqdm import TQDMNotebookCallback # keras, model definition... model.fit(X_train, Y_train, verbose=0, callbacks=[TQDMNotebookCallback()])
For plain text mode (e.g. for Python run from command line)
from keras_tqdm import TQDMCallback # keras, model definition... model.fit(X_train, Y_train, verbose=0, callbacks=[TQDMCallback()])
Advanced usage
Use keras_tqdm to utilize TQDM progress bars for Keras fit loops. keras_tqdm loops can be nested inside TQDM loops to display nested progress bars (although you can use them inside ordinary for loops as well). Set verbose=0 to suppress the default progress bar.
from keras_tqdm import TQDMCallback
from tqdm import tqdm
for model in tqdm(models, desc="Training several models"):
model.fit(x, y, verbose=0, callbacks=[TQDMCallback()])
For IPython and Jupyter notebook TQDMNotebookCallback instead of TQDMCallback. Use tqdm_notebook in your own code instead of tqdm. Formatting is controlled by python format strings. The default metric_format is "{name}: {value:0.3f}". For example, use TQDMCallback(metric_format="{name}: {value:0.6f}") for 6 decimal points or {name}: {value:e} for scientific notation.
Questions?
Please feel free to submit PRs and issues. Comments, questions, and requests are welcome. If you need more control, subclass TQDMCallback and override the tqdm function.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file keras_tqdm-2.0.0.tar.gz.
File metadata
- Download URL: keras_tqdm-2.0.0.tar.gz
- Upload date:
- Size: 4.4 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
dca24d533cd4a2e01938a7e780dd2f276cc0378193db22931e97d31cb6470d88
|
|
| MD5 |
2df1f1361d9bf93734d1d415a2657207
|
|
| BLAKE2b-256 |
70cd095d5359b9bd41c673aa7983b8c37ae6f8fd6e77df09f54eb33e38befea8
|
File details
Details for the file keras_tqdm-2.0.0-py2.py3-none-any.whl.
File metadata
- Download URL: keras_tqdm-2.0.0-py2.py3-none-any.whl
- Upload date:
- Size: 7.1 kB
- Tags: Python 2, Python 3
- Uploaded using Trusted Publishing? No
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
48841c97e6e6efd0a92676db9c43a8f669f74cd56cdd249b2125cb2726287a6a
|
|
| MD5 |
d0393111f495756bb9b650da90940422
|
|
| BLAKE2b-256 |
38cba47a8837236071b3002453b8e88bdea3ce70c2ed542b61686f61f2ea5f0e
|










