LIANA+: a one-stop-shop framework for cell-cell communication
Project description
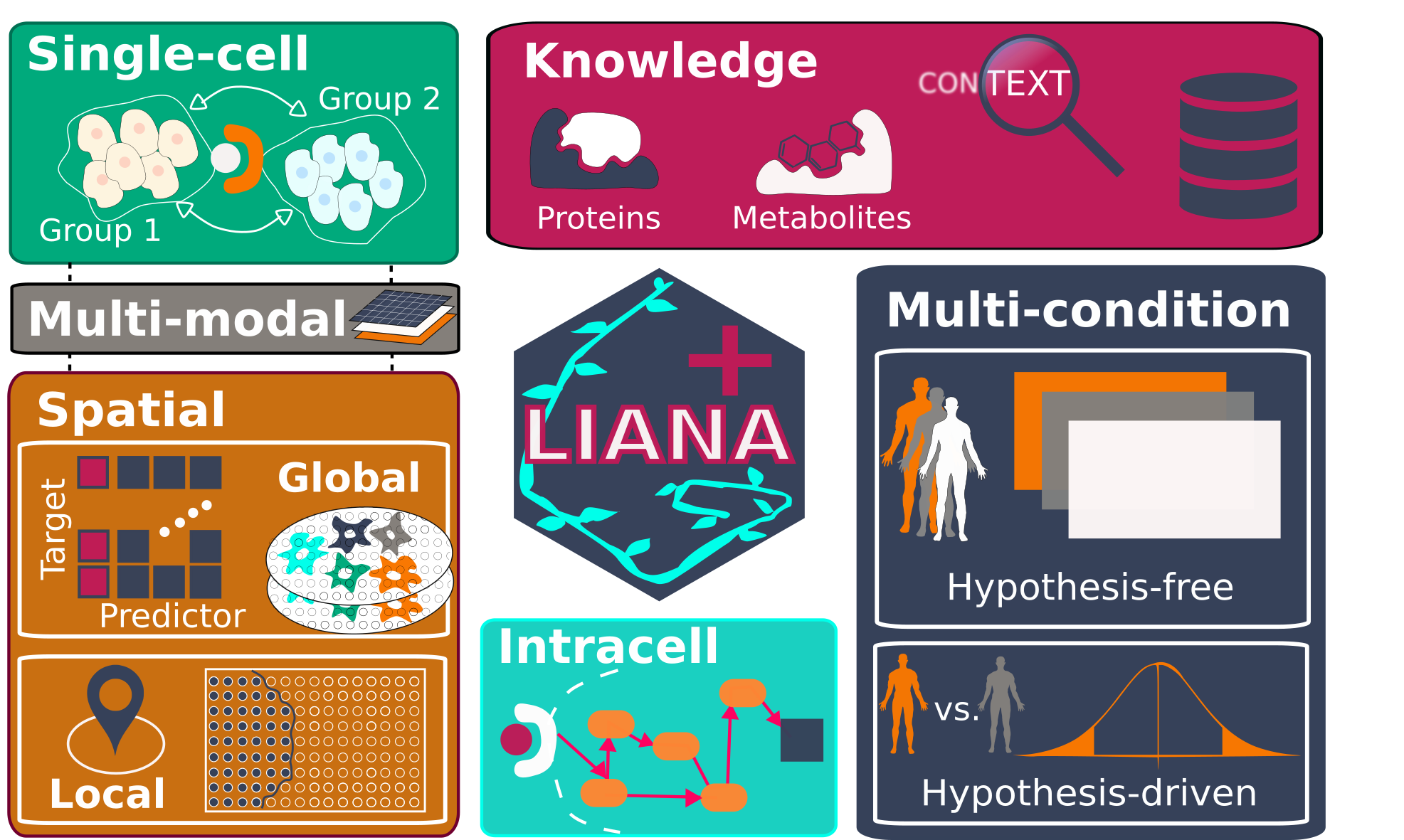
LIANA+: an all-in-one cell-cell communication framework 
LIANA+ is a scalable framework that adapts and extends existing methods and knowledge to study cell-cell communication in single-cell, spatially-resolved, and multi-modal omics data. It is part of the scverse ecosystem, and relies on AnnData & MuData objects as input.
Contributions
We welcome suggestions, ideas, and contributions! Please do not hesitate to contact us, open issues, and check the contributions guide.
Vignettes
A set of extensive vignettes can be found in the LIANA+ documentation.
Decision Tree
flowchart TD
Start[What type of data?] --> Spatial{Spatial<br/>coordinates?}
Start --> Modal{Multi-modal?}
%% Spatial branch
Spatial -->|Yes| SpatialRes{Resolution?}
SpatialRes -->|Single-cell| Inflow[Inflow Score]
SpatialRes -->|Spot-based| SpatialType{Analysis type?}
SpatialType -->|Bivariate| LocalQ{Local<br/>interactions?}
LocalQ -->|Yes| Local[Local Bivariate Metrics]
LocalQ -->|No| Global[Global Bivariate Metrics]
SpatialType -->|Unsupervised| MISTy[Multi-view Learning]
%% Non-spatial branch
Spatial -->|No| Compare{Compare across<br/>samples?}
Compare -->|Yes| Contrast{Specific<br/>contrast?}
Contrast -->|Yes| Targeted[Differential Contrasts]
Contrast -->|No| MOFA[MOFA+]
Contrast -->|No| Tensor[Tensor-cell2cell]
Tensor --> TensorExt[Extended Tutorials]
Compare -->|No| Steady[Steady-state LR Inference]
%% Multi-modal branch
Modal -->|Spatial| SMA[Multi-Modal Spatial]
Modal -->|Non-Spatial| SCMulti[Multi-Modal Single-Cell]
%% Metabolite sub-branch
SCMulti --> Metab[Metabolite-mediated CCC]
%% Links (click events)
click Inflow "https://liana-py.readthedocs.io/en/latest/notebooks/inflow_score.html"
click Local "https://liana-py.readthedocs.io/en/latest/notebooks/bivariate.html"
click Global "https://liana-py.readthedocs.io/en/latest/notebooks/bivariate.html"
click MISTy "https://liana-py.readthedocs.io/en/latest/notebooks/misty.html"
click Targeted "https://liana-py.readthedocs.io/en/latest/notebooks/targeted.html"
click MOFA "https://liana-py.readthedocs.io/en/latest/notebooks/mofatalk.html"
click Tensor "https://liana-py.readthedocs.io/en/latest/notebooks/liana_c2c.html"
click TensorExt "https://ccc-protocols.readthedocs.io/en/latest/"
click Steady "https://liana-py.readthedocs.io/en/latest/notebooks/basic_usage.html"
click SMA "https://liana-py.readthedocs.io/en/latest/notebooks/sma.html"
click SCMulti "https://liana-py.readthedocs.io/en/latest/notebooks/sc_multi.html"
click Metab "https://liana-py.readthedocs.io/en/latest/notebooks/sc_multi.html#metabolite-mediated-ccc-from-transcriptomics-data"
API
For further information please check LIANA's API documentation.
Cite LIANA+:
Dimitrov D., Schäfer P.S.L, Farr E., Rodriguez Mier P., Lobentanzer S., Badia-i-Mompel P., Dugourd A., Tanevski J., Ramirez Flores R.O. and Saez-Rodriguez J. LIANA+ provides an all-in-one framework for cell–cell communication inference. Nat Cell Biol (2024). https://doi.org/10.1038/s41556-024-01469-w
Dimitrov, D., Türei, D., Garrido-Rodriguez M., Burmedi P.L., Nagai, J.S., Boys, C., Flores, R.O.R., Kim, H., Szalai, B., Costa, I.G., Valdeolivas, A., Dugourd, A. and Saez-Rodriguez, J. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat Commun 13, 3224 (2022). https://doi.org/10.1038/s41467-022-30755-0
Please also consider citing any of the methods and/or resources that were particularly relevant for your research!
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file liana-1.7.1.tar.gz.
File metadata
- Download URL: liana-1.7.1.tar.gz
- Upload date:
- Size: 33.0 MB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
85cc3bf19e7562eb76d396e8318cf13ec884998b15188791aadfce9cadd598dd
|
|
| MD5 |
b6cc2338eb9ec38d54acad289150028b
|
|
| BLAKE2b-256 |
6c3e48b01fe2897cee8fae6dc44a38b9b18071a36847dc6b44dc01f67a778853
|
Provenance
The following attestation bundles were made for liana-1.7.1.tar.gz:
Publisher:
release.yaml on saezlab/liana-py
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
liana-1.7.1.tar.gz -
Subject digest:
85cc3bf19e7562eb76d396e8318cf13ec884998b15188791aadfce9cadd598dd - Sigstore transparency entry: 849875570
- Sigstore integration time:
-
Permalink:
saezlab/liana-py@276fc05d065006680940f1611e33aa9f04a683c4 -
Branch / Tag:
refs/tags/1.7.1 - Owner: https://github.com/saezlab
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
release.yaml@276fc05d065006680940f1611e33aa9f04a683c4 -
Trigger Event:
release
-
Statement type:
File details
Details for the file liana-1.7.1-py3-none-any.whl.
File metadata
- Download URL: liana-1.7.1-py3-none-any.whl
- Upload date:
- Size: 558.3 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.13.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
f4eead6df3f7fd07648fda77a50b50673270e1fca19030a7af802d57b84eb2e6
|
|
| MD5 |
fae2b271b77e508741768caab33eb987
|
|
| BLAKE2b-256 |
113524bcb5134ad91fb9787c85c0c1ef5f81b1b87adf9dfb29e8edb5e0050ca2
|
Provenance
The following attestation bundles were made for liana-1.7.1-py3-none-any.whl:
Publisher:
release.yaml on saezlab/liana-py
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
liana-1.7.1-py3-none-any.whl -
Subject digest:
f4eead6df3f7fd07648fda77a50b50673270e1fca19030a7af802d57b84eb2e6 - Sigstore transparency entry: 849875574
- Sigstore integration time:
-
Permalink:
saezlab/liana-py@276fc05d065006680940f1611e33aa9f04a683c4 -
Branch / Tag:
refs/tags/1.7.1 - Owner: https://github.com/saezlab
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
release.yaml@276fc05d065006680940f1611e33aa9f04a683c4 -
Trigger Event:
release
-
Statement type:
















