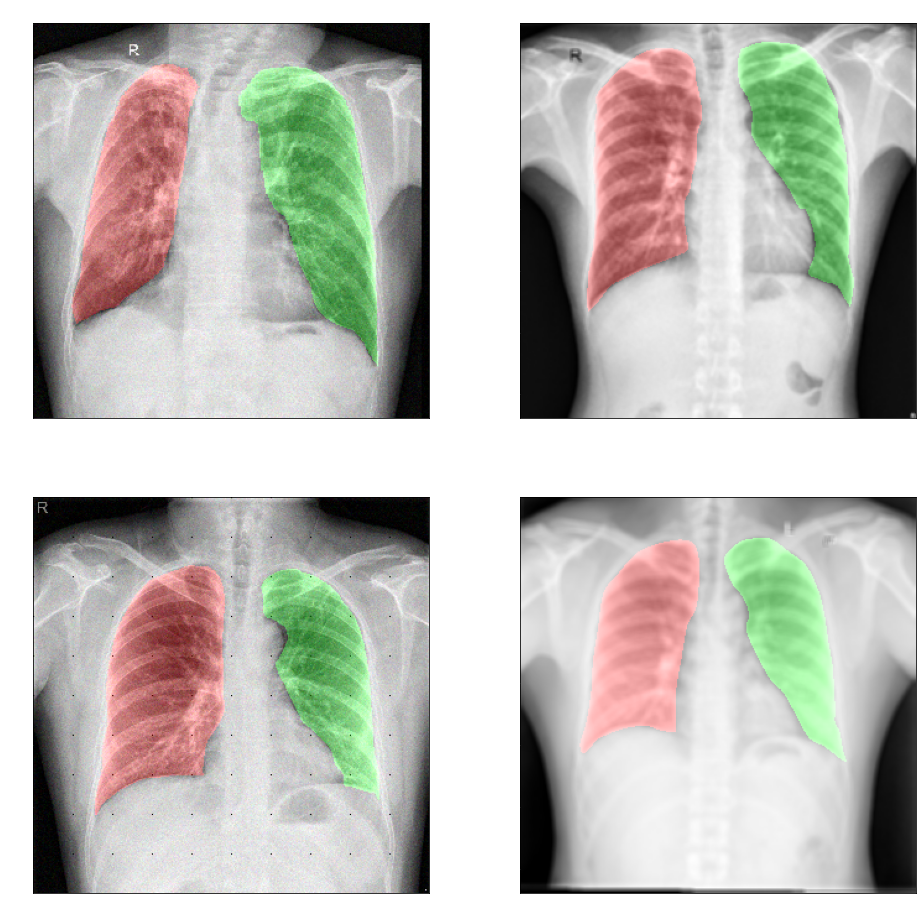
High quality model for lungs segmentation.
Project description
lungs_segmentation
Automated lung segmentation in chest-x ray
Train pipeline: https://github.com/alimbekovKZ/lungs_segmentation_train
Installation
pip install lungs-segmentation
Example inference
Jupyter notebook with the example:
WebApp
https://lungssegmentation.herokuapp.com/
Models weights
| model | best dice | Mb |
|---|---|---|
| resnet34 | 0.9657 | 103.4 |
| densenet121 | 0.9655 | 62.8 |
Usage
Code example for resnet34:
from lungs_segmentation.pre_trained_models import create_model
import lungs_segmentation.inference as inference
model = create_model("resnet34")
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
model = model.to(device)
plt.figure(figsize=(20,40))
plt.subplot(1,1,1)
image, mask = inference.inference(model,'739px-Chest_Xray_PA_3-8-2010.png', 0.2)
plt.imshow(inference.img_with_masks( image, [mask[0], mask[1]], alpha = 0.1))
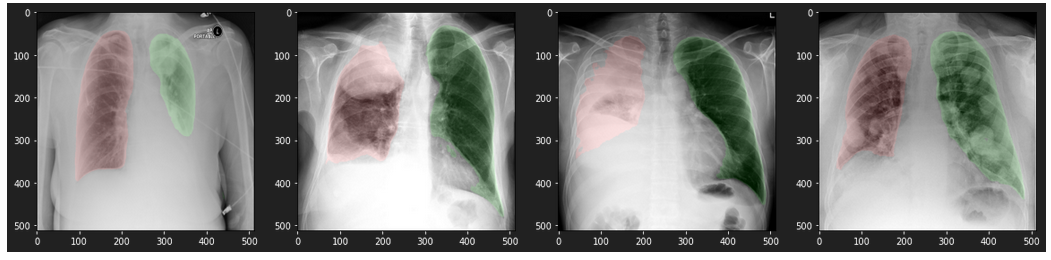
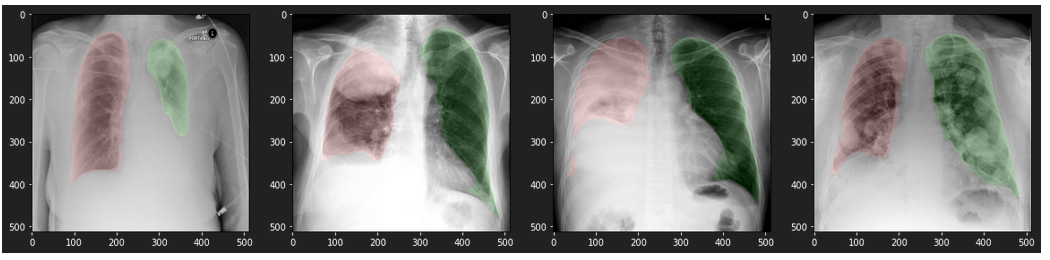
Results on data from the Internet
resnet34
densenet121
Authors
Renat Alimbekov, Ivan Vassilenko, Abylaikhan Turlassov
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file lungs_segmentation-1.1.2.tar.gz.
File metadata
- Download URL: lungs_segmentation-1.1.2.tar.gz
- Upload date:
- Size: 15.1 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.3.0 pkginfo/1.5.0.1 requests/2.21.0 setuptools/49.2.0 requests-toolbelt/0.9.1 tqdm/4.28.1 CPython/3.7.3
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
5406eafce99a60a938ab7ad900148508dac51ba269cefc938311ef814a57064f
|
|
| MD5 |
0f0ccf07efdbfe2ffe3eab2627d47d8c
|
|
| BLAKE2b-256 |
8057b7c4aae8ed7b19ce37bf3223f608bcf323296e618e8d2c1de3f376eaaed0
|
File details
Details for the file lungs_segmentation-1.1.2-py2.py3-none-any.whl.
File metadata
- Download URL: lungs_segmentation-1.1.2-py2.py3-none-any.whl
- Upload date:
- Size: 16.5 kB
- Tags: Python 2, Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.3.0 pkginfo/1.5.0.1 requests/2.21.0 setuptools/49.2.0 requests-toolbelt/0.9.1 tqdm/4.28.1 CPython/3.7.3
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
890b8c0c3fd37d2b3e7ada13bc2fda5955ece4c004691d99cb3451fe4e527b06
|
|
| MD5 |
ca872c89ecac23933754d2e90cd0a3a4
|
|
| BLAKE2b-256 |
6fd7623a222151a50c2b3034c4a25fb036355b0959bdbb95e44e638dc90614e3
|