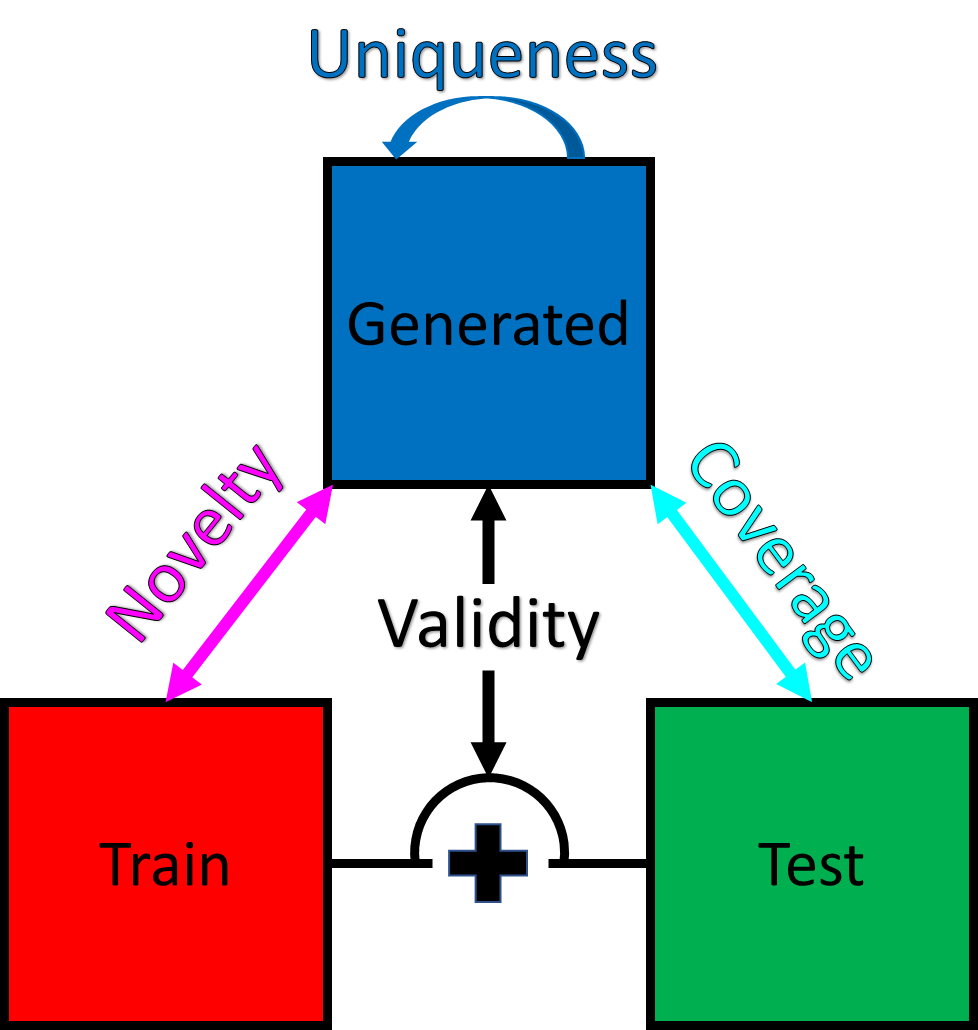
Generative materials benchmarking metrics, inspired by CDVAE.
Project description
NOTE: This is a WIP repository (as of 2022-08-06) being developed in parallel with
xtal2pngandmp-time-split. Feedback and contributions welcome!
matbench-genmetrics
Generative materials benchmarking metrics, inspired by guacamol and CDVAE.
This repository provides standardized benchmarks for benchmarking generative models for crystal structure. Each benchmark has a fixed dataset, a predefined split, and a notion of best (i.e. metric) associated with it.
Getting Started
Installation, a dummy example, output metrics for the example, and descriptions of the benchmark metrics.
Installation
Create a conda environment with the matbench-genmetrics package installed from the
conda-forge channel. Then activate the environment.
**NOTE: not available on conda-forge as of 2022-07-30, recipe under review by conda-forge team. So use
pip install matbench-genmetricsfor now
conda create --name matbench-genmetrics --channel conda-forge python==3.9.* matbench-genmetrics
conda activate matbench-genmetrics
NOTE: It doesn't have to be Python 3.9; you can remove
python==3.9.*altogether or change this to e.g.python==3.8.*. See Advanced Installation
Example
NOTE: be sure to set
dummy=Falsefor the real/full benchmark run. MPTSMetrics10 is intended for fast prototyping and debugging, as it assumes only 10 generated structures.
>>> from tqdm import tqdm
>>> from mp_time_split.utils.gen import DummyGenerator
>>> from matbench_genmetrics.core import MPTSMetrics10, MPTSMetrics100, MPTSMetrics1000, MPTSMetrics10000
>>> mptm = MPTSMetrics10(dummy=True)
>>> for fold in mptm.folds:
>>> train_val_inputs = mptm.get_train_and_val_data(fold)
>>> dg = DummyGenerator()
>>> dg.fit(train_val_inputs)
>>> gen_structures = dg.gen(n=mptm.num_gen)
>>> mptm.record(fold, gen_structures)
Output
print(mptm.recorded_metrics)
{
0: {
"validity": 0.4375,
"coverage": 0.0,
"novelty": 1.0,
"uniqueness": 0.9777777777777777,
},
1: {
"validity": 0.4390681003584229,
"coverage": 0.0,
"novelty": 1.0,
"uniqueness": 0.9333333333333333,
},
2: {
"validity": 0.4401197604790419,
"coverage": 0.0,
"novelty": 1.0,
"uniqueness": 0.8222222222222222,
},
3: {
"validity": 0.4408740359897172,
"coverage": 0.0,
"novelty": 1.0,
"uniqueness": 0.8444444444444444,
},
4: {
"validity": 0.4414414414414415,
"coverage": 0.0,
"novelty": 1.0,
"uniqueness": 0.9111111111111111,
},
}
Metrics
| Metric | Description |
|---|---|
| Validity | One minus (Wasserstein distance between distribution of space group numbers for train and generated structures divided by distance of dummy case between train and space_group_number == 1). See also https://github.com/sparks-baird/matbench-genmetrics/issues/44 |
| Coverage | Match counts between held-out test structures and generated structures divided by number of test structures ("predict the future"). |
| Novelty | One minus (match counts between train structures and generated structures divided by number of generated structures). |
| Uniqueness | One minus (non-self-comparing match counts within generated structures divided by total possible non-self-comparing matches). |
A match is when StructureMatcher(stol=0.5, ltol=0.3, angle_tol=10.0).fit(s1, s2)
evaluates to True.
Advanced Installation
Anaconda (conda) installation (recommended)
(2022-07-30, conda-forge installation pending, fallback to pip install xtal2png as separate command)
Create and activate a new conda environment named xtal2png (-n) that will search for and install the xtal2png package from the conda-forge Anaconda channel (-c).
conda env create -n xtal2png -c conda-forge xtal2png
conda activate xtal2png
Alternatively, in an already activated environment:
conda install -c conda-forge xtal2png
If you run into conflicts with packages you are integrating with xtal2png, please try installing all packages in a single line of code (or two if mixing conda and pip packages in the same environment) and installing with mamba (source).
PyPI (pip) installation
Create and activate a new conda environment named matbench-genmetrics (-n) with python==3.9.* or your preferred Python version, then install matbench-genmetrics via pip.
conda create -n xtal2png python==3.9.*
conda activate xtal2png
pip install xtal2png
Editable installation
In order to set up the necessary environment:
-
clone and enter the repository via:
git clone https://github.com/sparks-baird/matbench-genmetrics.git cd matbench-genmetrics
-
create and activate a new conda environment (optional, but recommended)
conda env create --name matbench-genmetrics python==3.9.* conda activate matbench-genmetrics
-
perform an editable (
-e) installation in the current directory (.):pip install -e .
NOTE: Some changes, e.g. in
setup.cfg, might require you to runpip install -e .again.
Optional and needed only once after git clone:
-
install several pre-commit git hooks with:
pre-commit install # You might also want to run `pre-commit autoupdate`
and checkout the configuration under
.pre-commit-config.yaml. The-n, --no-verifyflag ofgit commitcan be used to deactivate pre-commit hooks temporarily. -
install nbstripout git hooks to remove the output cells of committed notebooks with:
nbstripout --install --attributes notebooks/.gitattributes
This is useful to avoid large diffs due to plots in your notebooks. A simple
nbstripout --uninstallwill revert these changes.
Then take a look into the scripts and notebooks folders.
Dependency Management & Reproducibility
- Always keep your abstract (unpinned) dependencies updated in
environment.ymland eventually insetup.cfgif you want to ship and install your package viapiplater on. - Create concrete dependencies as
environment.lock.ymlfor the exact reproduction of your environment with:conda env export -n matbench-genmetrics -f environment.lock.yml
For multi-OS development, consider using--no-buildsduring the export. - Update your current environment with respect to a new
environment.lock.ymlusing:conda env update -f environment.lock.yml --prune
Project Organization
├── AUTHORS.md <- List of developers and maintainers.
├── CHANGELOG.md <- Changelog to keep track of new features and fixes.
├── CONTRIBUTING.md <- Guidelines for contributing to this project.
├── Dockerfile <- Build a docker container with `docker build .`.
├── LICENSE.txt <- License as chosen on the command-line.
├── README.md <- The top-level README for developers.
├── configs <- Directory for configurations of model & application.
├── data
│ ├── external <- Data from third party sources.
│ ├── interim <- Intermediate data that has been transformed.
│ ├── processed <- The final, canonical data sets for modeling.
│ └── raw <- The original, immutable data dump.
├── docs <- Directory for Sphinx documentation in rst or md.
├── environment.yml <- The conda environment file for reproducibility.
├── models <- Trained and serialized models, model predictions,
│ or model summaries.
├── notebooks <- Jupyter notebooks. Naming convention is a number (for
│ ordering), the creator's initials and a description,
│ e.g. `1.0-fw-initial-data-exploration`.
├── pyproject.toml <- Build configuration. Don't change! Use `pip install -e .`
│ to install for development or to build `tox -e build`.
├── references <- Data dictionaries, manuals, and all other materials.
├── reports <- Generated analysis as HTML, PDF, LaTeX, etc.
│ └── figures <- Generated plots and figures for reports.
├── scripts <- Analysis and production scripts which import the
│ actual PYTHON_PKG, e.g. train_model.
├── setup.cfg <- Declarative configuration of your project.
├── setup.py <- [DEPRECATED] Use `python setup.py develop` to install for
│ development or `python setup.py bdist_wheel` to build.
├── src
│ └── matbench_genmetrics <- Actual Python package where the main functionality goes.
├── tests <- Unit tests which can be run with `pytest`.
├── .coveragerc <- Configuration for coverage reports of unit tests.
├── .isort.cfg <- Configuration for git hook that sorts imports.
└── .pre-commit-config.yaml <- Configuration of pre-commit git hooks.
Note
This project has been set up using PyScaffold 4.2.2.post1.dev2+ge50b5e1 and the dsproject extension 0.7.2.post1.dev2+geb5d6b6.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for matbench-genmetrics-0.5.2.tar.gz
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | acc7fdf45addc4715514a4ce0131f1098fcb9fe6d6dbc081b12afa2c8dce1b24 |
|
| MD5 | 467e70cbfa9f21fad5c6efcc6471cd27 |
|
| BLAKE2b-256 | 07cd8fb65cc94275d800ef2f3f73804bac7c3505295de03c9c14972ff5403b76 |
Hashes for matbench_genmetrics-0.5.2-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 141e8697d126779d406b439409592051135bb8250a155786e699b421ff557843 |
|
| MD5 | 907bae2f8050c3076f6d6e0af95be864 |
|
| BLAKE2b-256 | 27568ae0f29a3ed95c0c4fb563ede192af22b4d9920c5788d1a2960c7f2b7116 |