Python-based Illumina methylation array processing and analysis software composite package
Project description
Overview
This meta package will install the python packages that together make up the methylsuite:
methylprepis a python package for processing DNA methylation data from Illumina arrays or downloading GEO datasets from NIH. It provides:- Support for Illumina arrays (27k, 450k, EPIC, mouse).
- Support for analyzing public data sets from GEO (the NIH Gene Expression Omnibus is a database).
- Support for managing data in
PandasDataFrames. - data cleaning functions during processing, including:
- infer type-I channel switch
- NOOB
- poobah (p-value with out-of-band array hybridization, for filtering lose signal-to-noise probes)
- qualityMask (to exclude historically less reliable probes)
- nonlinear dye bias correction (AKA signal quantile normalization between red/green channels across a sample)
- calculate beta-value, m-value, or copy-number matrix
- large batch memory management, by splitting it up into smaller batches during processing
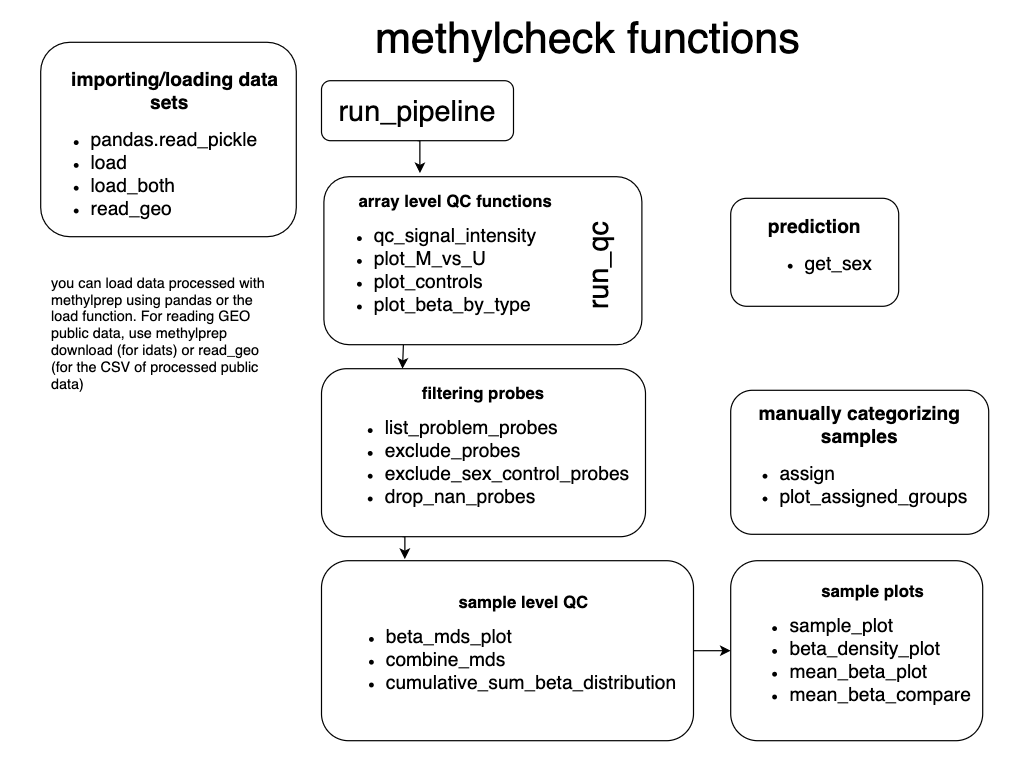
methylcheckincludes:- quality control (QC) functions for filtering out unreliable probes, based on the published literature, sample outlier detection, plots similar to Genome Studio functions, sex prediction, and and interactive method for assigning samples to groups, based on array data, in a Jupyter notebook.
methylizeprovides these analysis functions:- differentially methylated probe statistics (between treatment and control samples).
- volcano plots (which probes are the most different).
- manhattan plot (where in genome are the differences).
Parts of methylsuite
Data Processing
Quality Control
Installation
This all-in-one command will install all three packages and their required dependencies.
pip3 install methylsuite
This command was tested in a naive minimal conda environment, created thusly:
conda create -n testsuite
Afterwards, I had to install statsmodels using conda apart from the other dependencies, like so:
conda install statsmodels
pip3 install methylsuite
(Statsmodels uses additional C-compilers during installation)
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file methylsuite-1.4.0.tar.gz.
File metadata
- Download URL: methylsuite-1.4.0.tar.gz
- Upload date:
- Size: 2.9 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.1 importlib_metadata/3.10.0 pkginfo/1.7.0 requests/2.27.1 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.8.8
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
a0954d9d3859bad30a0775291ed8cb5a9f9787e32d50b4f108675d24909db79e
|
|
| MD5 |
9c5cf4e4474bd12668c1760d9455317b
|
|
| BLAKE2b-256 |
dfc181186c53b4bf8ac5cf6766ceaea7a39aef00ad1da3b486cddac81837bca3
|
File details
Details for the file methylsuite-1.4.0-py3-none-any.whl.
File metadata
- Download URL: methylsuite-1.4.0-py3-none-any.whl
- Upload date:
- Size: 2.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.1 importlib_metadata/3.10.0 pkginfo/1.7.0 requests/2.27.1 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.8.8
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
3ebed68be5faf79ce78168a6cd6a164da28aba955fe0a3d7497f921fefceee0b
|
|
| MD5 |
9fe06e66a69063b6f60654f02b5c62f8
|
|
| BLAKE2b-256 |
e05c9bdb3d00e831955e33fe98df6f0bd2a330609bd0c36f52e5e0dad522963f
|