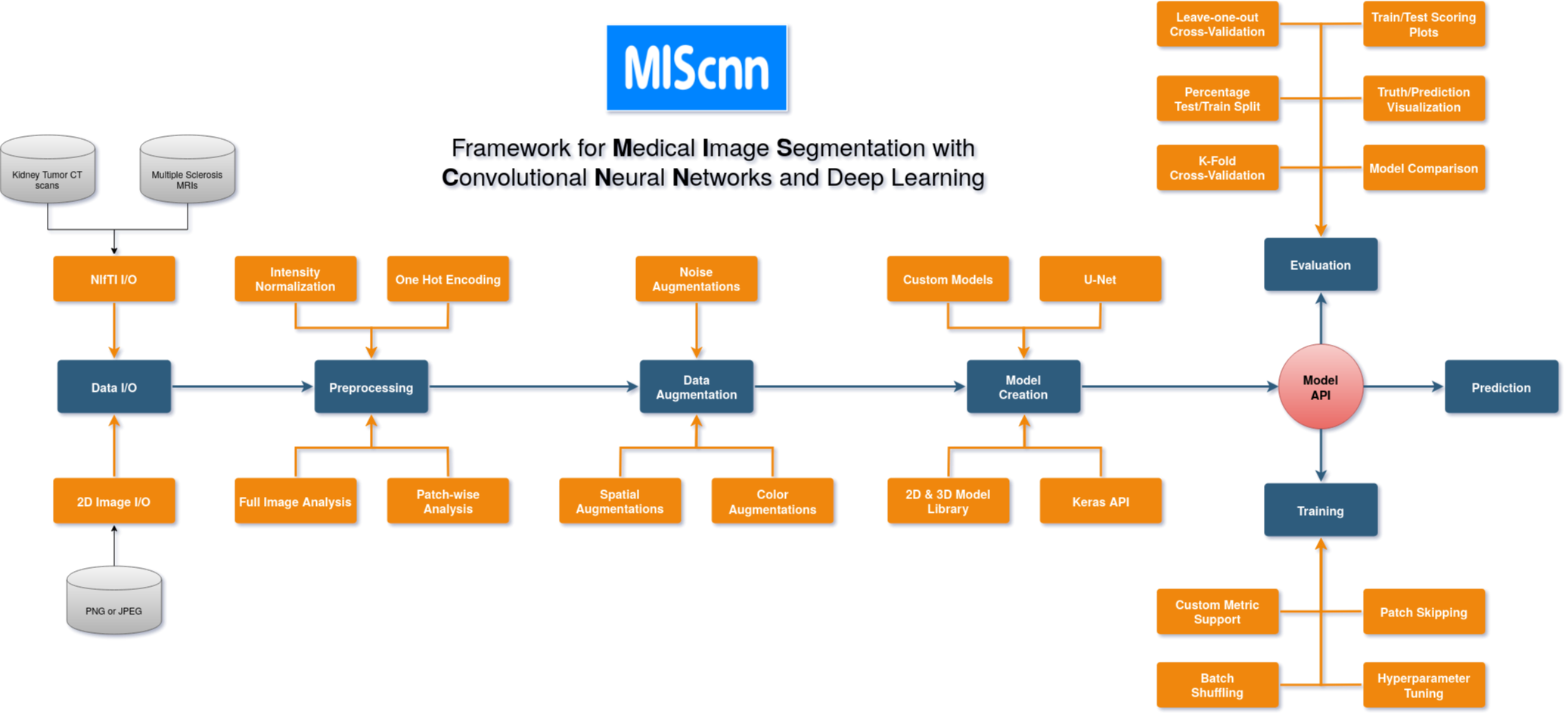
Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning
Project description
MIScnn: Medical Image Segmentation with Convolutional Neural Networks
The open-source Python library MIScnn is an intuitive API allowing fast setup of medical image segmentation pipelines with state-of-the-art convolutional neural network and deep learning models in just a few lines of code.
MIScnn provides several core features:
- 2D/3D medical image segmentation for binary and multi-class problems
- Data I/O, preprocessing and data augmentation for biomedical images
- Patch-wise and full image analysis
- State-of-the-art deep learning model and metric library
- Intuitive and fast model utilization (training, prediction)
- Multiple automatic evaluation techniques (e.g. cross-validation)
- Custom model, data I/O, pre-/postprocessing and metric support
- Based on Keras with Tensorflow as backend
Getting started: 30 seconds to a MIS pipeline
Create a Data I/O instance with an already provided interface for your specific data format.
from miscnn.data_loading.data_io import Data_IO
from miscnn.data_loading.interfaces.nifti_io import NIFTI_interface
# Create an interface for kidney tumor CT scans in NIfTI format
interface = NIFTI_interface(pattern="case_0000[0-2]", channels=1, classes=3)
# Initialize data path and create the Data I/O instance
data_path = "/home/mudomini/projects/KITS_challenge2019/kits19/data.original/"
data_io = Data_IO(interface, data_path)
Create a Preprocessor instance to configure how to preprocess the data into batches.
from miscnn.processing.preprocessor import Preprocessor
pp = Preprocessor(data_io, batch_size=4, analysis="patchwise-crop", patch_shape=(128,128,128))
Create a deep learning neural network model with a standard U-Net architecture.
from miscnn.neural_network.model import Neural_Network
from miscnn.neural_network.architecture.unet.standard import Architecture
unet_standard = Architecture()
model = Neural_Network(preprocessor=pp, architecture=unet_standard)
Congratulations to your ready-to-use Medical Image Segmentation pipeline including data I/O, preprocessing and data augmentation with default setting.
Let's run a model training on our data set. Afterwards, predict the segmentation of a sample using the fitted model.
# Training the model with all except one sample for 500 epochs
sample_list = data_io.get_indiceslist()
model.train(sample_list[0:-1], epochs=500)
# Predict the one remaining sample
pred = model.predict([sample_list[-1]], direct_output=True)
Now, let's run a 5-fold Cross-Validation with our model, create automatically evaluation figures and save the results into the directory "evaluation_results".
from miscnn.evaluation.cross_validation import cross_validation
cross_validation(sample_list, model, k_fold=5, epochs=100,
evaluation_path="evaluation_results", draw_figures=True)
Installation
There are two ways to install MIScnn:
- Install MIScnn from PyPI (recommended):
Note: These installation steps assume that you are on a Linux or Mac environment. If you are on Windows or in a virtual environment without root, you will need to remove sudo to run the commands below.
sudo pip install miscnn
- Alternatively: install MIScnn from the GitHub source:
First, clone MIScnn using git:
git clone https://github.com/frankkramer-lab/MIScnn.git
Then, cd to the MIScnn folder and run the install command:
cd MIScnn
sudo python setup.py install
Author
Dominik Müller
Email: dominik.mueller@informatik.uni-augsburg.de
IT-Infrastructure for Translational Medical Research
University Augsburg
Bavaria, Germany
How to cite / More information
Dominik Müller and Frank Kramer. (2019)
MIScnn: A Framework for Medical Image Segmentation with Convolutional Neural Networks and Deep Learning.
License
This project is licensed under the GNU GENERAL PUBLIC LICENSE Version 3.
See the LICENSE.md file for license rights and limitations.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file miscnn-0.14.tar.gz.
File metadata
- Download URL: miscnn-0.14.tar.gz
- Upload date:
- Size: 36.2 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.13.0 pkginfo/1.5.0.1 requests/2.22.0 setuptools/41.2.0 requests-toolbelt/0.9.1 tqdm/4.35.0 CPython/3.5.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
8cf5212d31f27a0844e97ccd04d4fe50e6132e270b06437147c0d5514c7ae6a1
|
|
| MD5 |
70791a659a8a3844e4f0f6dde9d44d40
|
|
| BLAKE2b-256 |
ac90b8829faab9eff12ef419f725019926c98a84303cc33cde5a940360edc459
|
File details
Details for the file miscnn-0.14-py3-none-any.whl.
File metadata
- Download URL: miscnn-0.14-py3-none-any.whl
- Upload date:
- Size: 72.6 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.13.0 pkginfo/1.5.0.1 requests/2.22.0 setuptools/41.2.0 requests-toolbelt/0.9.1 tqdm/4.35.0 CPython/3.5.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
8c7b3dac16c71911f7f4ba0889b590846278c7889b4b817f450f3912a2ca72c0
|
|
| MD5 |
c5046fcd9f3b3c150431af46312e5b87
|
|
| BLAKE2b-256 |
79b285186e2c8efe91bb3a56965cdd78d75197f9eb1986f57372866d3ec061e3
|