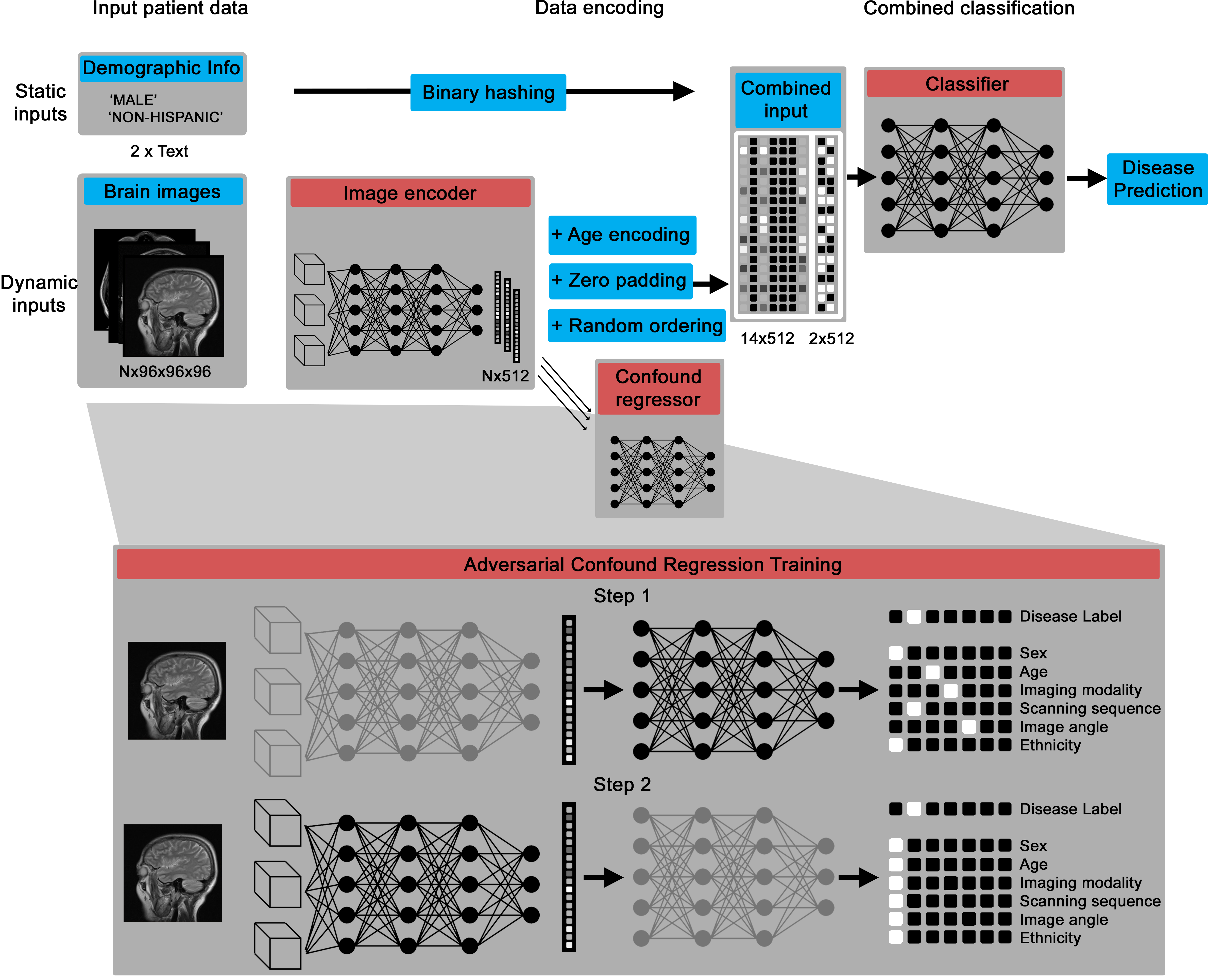
Deep learning library to encode multiple brain images and other electronic health record data in disease detection.
Project description
Multi-Input Medical Image Machine Learning Toolkit
The Multi-Input Medical Image Machine Learning Toolkit (MultiMedImageML) is a library of Pytorch functions that can encode multiple 3D images (designed specifically for brain images) and offer a single- or multi-label output, such as a disease detection.
Thus, with a dataset of brain images and labels, you can train a model to predict dementia or multiple sclerosis from multiple input brain images.
To install Multi Med Image ML, simply type into a standard UNIX terminal
pip install multi-med-image-ml
Overview
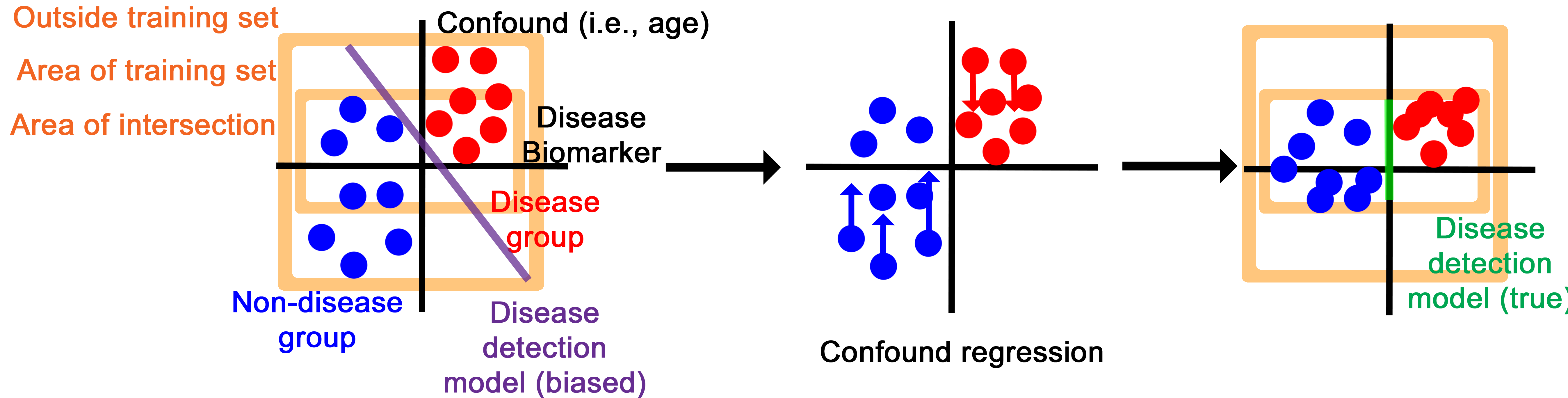
The core deep learning architecture is a Pytorch model that can take in variable numbers of 3D images (between one and 14 by default), then encodes them into a numerical vector and, through an adversarial training process, creates an intermediate representation that contains information about disease biomarkers but not confounds, like patient age and scanning site.
The confound regression process essentially disguises the intermediary representation to have disease biomarker features while imitating the confounding features of other groups.
Getting Started
See the Documentation.
Datasets
This may be used with either public benchmark datasets of brain images or internal hospital records, so long as they're represented as DICOM or NIFTI images. It was largely tested on ADNI and data internal to MGH. If they're represented as DICOM images, they are converted to NIFTI with metadata represented as a JSON file using dcm2niix. They may be further converted to NPY files, which are resized to a specific dimension, with the metadata represented in a pandas dataframe.
The MedImageLoader builds up this representation automatically, but it is space-intensive to do so.
Data may be represented with a folder structure.
.
└── control
├── 941_S_7051
│ ├── Axial_3TE_T2_STAR
│ │ └── 2022-03-07_11_03_03.0
│ │ ├── I1553008
│ │ │ ├── I1553008_Axial_3TE_T2_STAR_20220307110304_5_e3_ph.json
│ │ │ └── I1553008_Axial_3TE_T2_STAR_20220307110304_5_e3_ph.nii.gz
│ │ └── I1553014
│ │ ├── I1553014_Axial_3TE_T2_STAR_20220307110304_5_ph.json
│ │ └── I1553014_Axial_3TE_T2_STAR_20220307110304_5_ph.nii.gz
│ ├── HighResHippocampus
│ │ └── 2022-03-07_11_03_03.0
│ │ └── I1553013
│ │ ├── I1553013_HighResHippocampus_20220307110304_11.json
│ │ └── I1553013_HighResHippocampus_20220307110304_11.nii.gz
│ └── Sagittal_3D_FLAIR
│ └── 2022-03-07_11_03_03.0
│ └── I1553012
│ ├── I1553012_Sagittal_3D_FLAIR_20220307110304_3.json
│ └── I1553012_Sagittal_3D_FLAIR_20220307110304_3.nii.gz
└── 941_S_7087
├── Axial_3D_PASL__Eyes_Open_
│ └── 2022-06-15_14_38_03.0
│ └── I1591322
│ ├── I1591322_Axial_3D_PASL_(Eyes_Open)_20220615143803_6.json
│ └── I1591322_Axial_3D_PASL_(Eyes_Open)_20220615143803_6.nii.gz
└── Perfusion_Weighted
└── 2022-06-15_14_38_03.0
└── I1591323
├── I1591323_Axial_3D_PASL_(Eyes_Open)_20220615143803_7.json
└── I1591323_Axial_3D_PASL_(Eyes_Open)_20220615143803_7.nii.gz
In the case of the above folder structure, "/path/to/control" may simply be input into the MedImageLoader function. For multiple labels, "/path/to/test", "/path/to/test2", and so on, may also be input.
Labels and Confounds
MIMIM enables for the representation of labels to classify by and confounds to regress. Confounds are represented as strings and labels can be represented as either strings or the input folder structure to MedImageLoader.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file multi_med_image_ml-0.0.25.tar.gz.
File metadata
- Download URL: multi_med_image_ml-0.0.25.tar.gz
- Upload date:
- Size: 3.4 MB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/5.1.0 CPython/3.12.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 45397b3ecdbcd4e05801bf762dcebaf85bd1b11d7599b297db8d16ab45519f60 |
|
| MD5 | a52cc479b250084834072f6a536ec45c |
|
| BLAKE2b-256 | 9932fe9e38b045f28e9535bdb4212cef2d65842d5cb15879f25bb035d10694c9 |
File details
Details for the file multi_med_image_ml-0.0.25-py3-none-any.whl.
File metadata
- Download URL: multi_med_image_ml-0.0.25-py3-none-any.whl
- Upload date:
- Size: 51.2 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/5.1.0 CPython/3.12.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 97904ee83393c58a471705a25dfac7cb6c380eb70ab4ba3f81afe4b5e882a2a2 |
|
| MD5 | 8a6fe0c0eb793bfc59e456223d4ae9b9 |
|
| BLAKE2b-256 | 0936443c97eaf91208d9dc3e06990e9edc864267accda12705535e8addc92e26 |