A compilation of bioinformatic and computation biology inspired python tools
Project description

PURPOSE
Bring broadscale comparative genomics to the masses.
Mycotools is a compilation of computational biology tools and database MycotoolsDB software that facilitate large-scale prokaryotic and fungal comparative genomics. MycotoolsDB locally assimilates all NCBI and MycoCosm (Joint Genome Institute) genomes into a database schema with uniform file curation, scalability, and automation as guiding principles.
- Installation is as simple as
updateDB.py --init <DIR>. No more limited and incomplete databases. updateDB.py --updatebrings the database to the current date. No more database obsolecence in the wake of accelerating genome availability.- The MycotoolsDB (MTDB) uniformly curates the numerous notorious iterations of
the
gff, allowing for reliable analyses and format expectations from multiple eras. - The
.mtdbdatabase format enables swift transitions from analyses with datasets of 100,000s genomes to as few as a lineage of interest. No more scrambling to acquire files for an analysis.
Mycotools is currently available as a beta subset of the full suite, excluding the database assimilation tools. MycotoolsDB is currently restricted to Ohio Supercomputer Center - if you are interested in early access, please email konkelzach@protonmail.com.
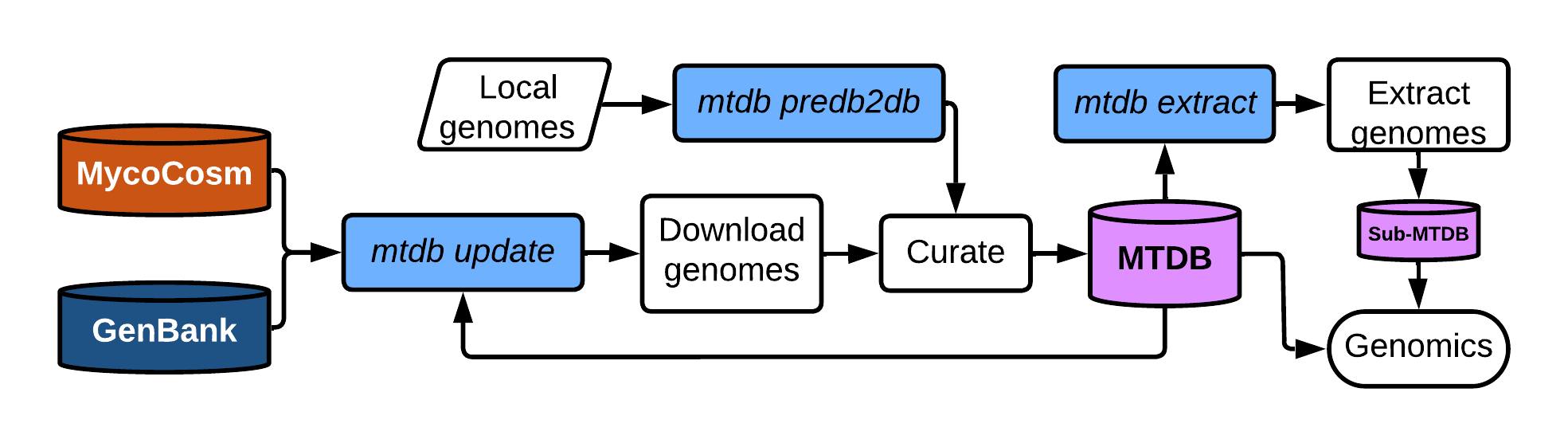
By integrating with the curated MycotoolsDB, Mycotools aids routine-complex tasks like retrieving gff or fasta accessions; running and compiling fastas of MycotoolsDB BLAST/hmmsearches; biology-based database manipulation tools; automated phylogenetic analysis pipelines from blast to Pfam extraction to tree prediction, etc etc. Mycotools includes sets of utilities that also enable easy acquisition of batches of sequence data using ncbiDwnld.py and jgiDwnld.py.
USAGE
Check out README.md for install and the USAGE.md for a guide.
CITING
If Mycotools contribute to your analysis, please cite this git repository (gitlab.com/xonq/mycotools) and mention the Mycotools version in line.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file mycotools-0.22.tar.gz.
File metadata
- Download URL: mycotools-0.22.tar.gz
- Upload date:
- Size: 98.7 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.3.0 pkginfo/1.7.0 requests/2.25.1 setuptools/49.6.0.post20210108 requests-toolbelt/0.9.1 tqdm/4.60.0 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
9f5bec618b5ba7846ffbe219289da1093681ff16882c01a8666a43b22f7b7728
|
|
| MD5 |
6e2c39c9e279c030c8e99654f250b8b5
|
|
| BLAKE2b-256 |
3b3e80268b597713a3d4b47a2bba2a01a0a145e072715cb20d7ce0dda780d12f
|
File details
Details for the file mycotools-0.22-py3-none-any.whl.
File metadata
- Download URL: mycotools-0.22-py3-none-any.whl
- Upload date:
- Size: 207.8 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.3.0 pkginfo/1.7.0 requests/2.25.1 setuptools/49.6.0.post20210108 requests-toolbelt/0.9.1 tqdm/4.60.0 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
65446eb8164e39ff28617e204fd0d0e09baa9ce2670f917e39f9a45066bc90b5
|
|
| MD5 |
f0a40dffa70f482e8b95a1e6427a9692
|
|
| BLAKE2b-256 |
03620a3a9ab93980e74b66a158c1880a3bbeed3f81819b5348363a5f0f35799b
|











