ncem. Learning cell communication from spatial graphs of cells.
Project description
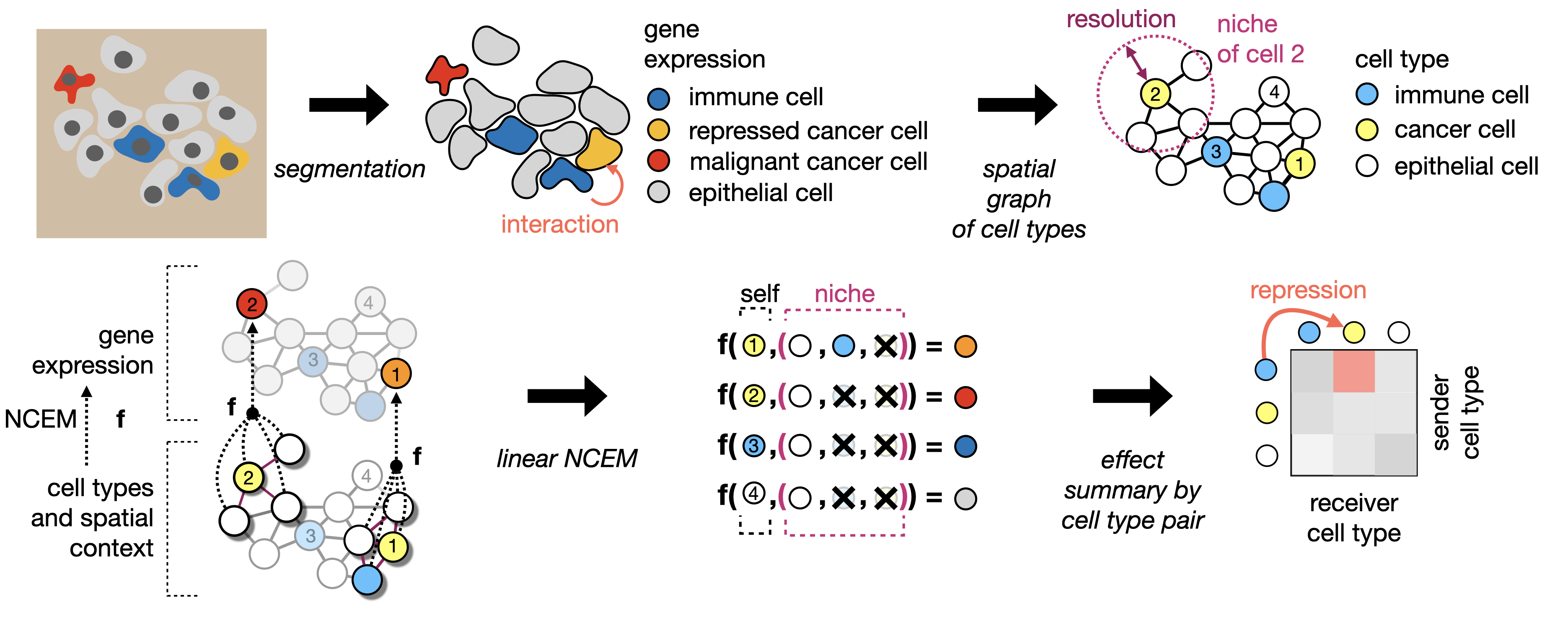
Features
ncem is a model repository in a single python package for the manuscript Fischer, D. S., Schaar, A. C. and Theis, F. Learning cell communication from spatial graphs of cells. 2021. (preprint)
Installation
You can install ncem via pip from PyPI:
$ pip install ncemCredits
This package was created with cookietemple using Cookiecutter based on Hypermodern_Python_Cookiecutter.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file ncem-0.1.5.tar.gz.
File metadata
- Download URL: ncem-0.1.5.tar.gz
- Upload date:
- Size: 105.6 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.17
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
34914d115040aec051eccfc30f510019d0bc78e54460aff5ddad3a8617c318df
|
|
| MD5 |
7b0761bfd050a0e0c5b82b9789d42fb7
|
|
| BLAKE2b-256 |
4be0bbc2791dfee03b7b9f831b43049e2156849e6e7823c74d50935d3bfa9506
|
File details
Details for the file ncem-0.1.5-py3-none-any.whl.
File metadata
- Download URL: ncem-0.1.5-py3-none-any.whl
- Upload date:
- Size: 143.6 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.17
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
496e2561b136512241e7f07fc0ca7b61698cd04d778be5e57cb7f4a533abd117
|
|
| MD5 |
79081f621803bebf51f1b0da3173b488
|
|
| BLAKE2b-256 |
0aa5f1684287d01936e794f3cdbfe3d52b360b8a81e304c84642d8891bdb38ee
|


















