A Python implementation of the OrthoANI algorithm for nucleotide identity measurement.
Project description
(Py)OrthoANI 
A Python implementation of the OrthoANI algorithm for nucleotide identity measurement.
🗺️ Overview
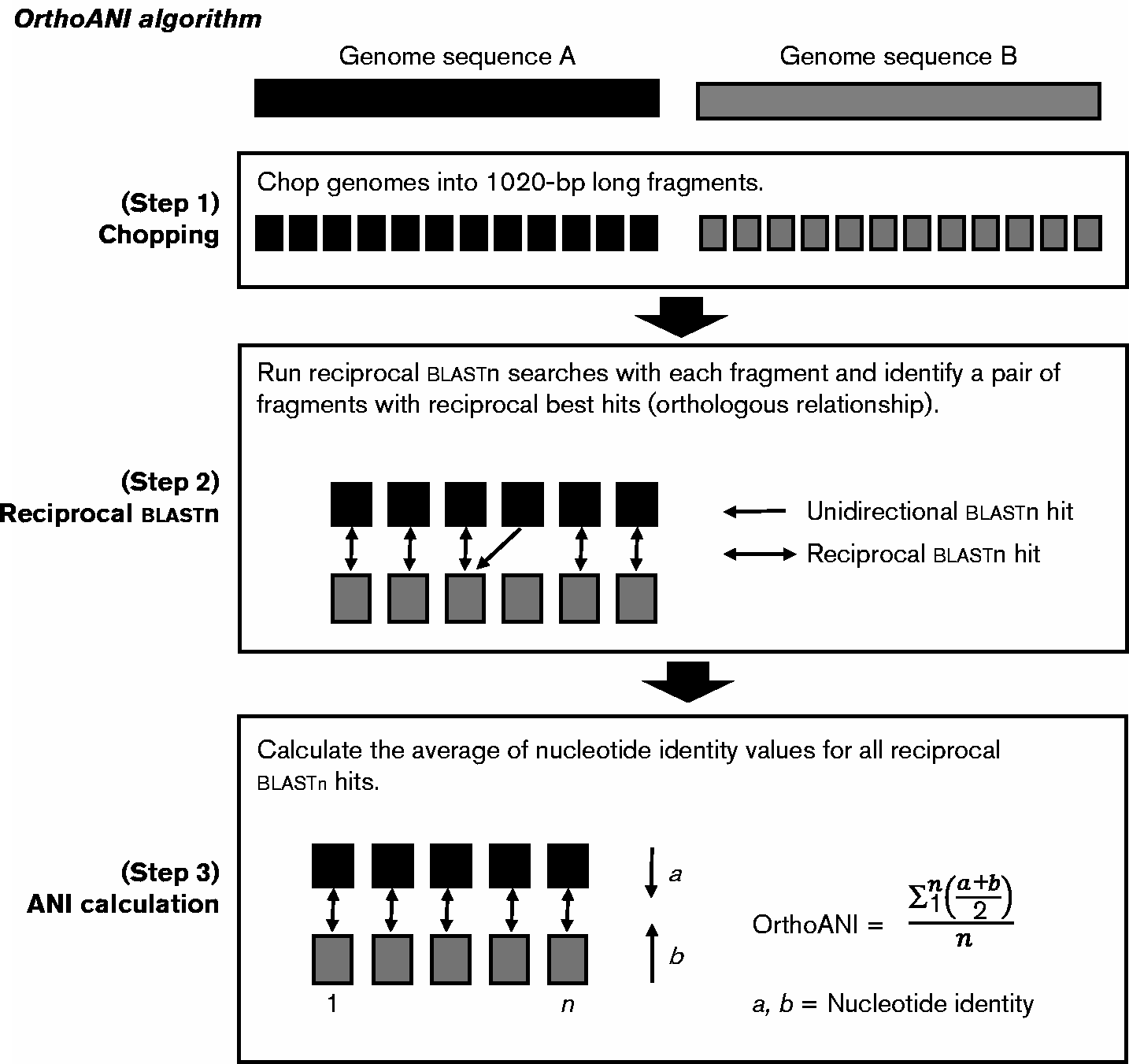
OrthoANI is a metric proposed by Lee et al.[1] in 2016 to improve computation of Average Nucleotide Identity. It uses BLASTn to find orthologous blocks in a pair of sequences, and then computes the average identity only considering alignments of reciprocal orthologs.
This project is a reimplementation of the closed-source Java implementation
provided by the authors on ezbiocloud.net.
It relies on Biopython to handle the I/O, and calls
the BLAST+ binaries using the subprocess module of the Python standard
library.
🔧 Installing
Installing with pip is the easiest:
$ pip install orthoani
orthoani also requires the BLAST+ binaries to be installed on your machine
and available somewhere in your $PATH.
💡 Example
Use Biopython to load two FASTA files, and then orthoani.orthoani to compute
the OrthoANI metric between them:
import orthoani
from Bio.SeqIO import read
genome_1 = read("sequence1.fa", "fasta")
genome_2 = read("sequence2.fa", "fasta")
ani = orthoani.orthoani(genome_1, genome_2)
orthoani can also be used from the CLI using a very simple command-line
interface:
$ orthoani -q sequence1.fa -r sequence2.fa
0.5725
🐏 Memory
orthoani uses the machine temporary folder to handle BLAST+ input and output
files, which is configurable through
tempfile.tempdir.
On some systems (like ArchLinux), this filesystem can reside in memory, which means
that your computer could have trouble processing very large files. If this
happens, try changing the value of the tempfile.tempdir to a directory that
is actually located on physical storage.
📏 Precision
Values computed by this package and the original Java implementation may differ slightly because in Java the authors perform rounding of floating-point values at the sub-percent level, while this library uses the full values.
📜 About
This library is provided under the open-source MIT license.
This project is in no way not affiliated, sponsored, or otherwise endorsed by the original OrthoANI authors. It was developed by Martin Larralde during his PhD project at the European Molecular Biology Laboratory in the Zeller team.
📚 References
- [1] Imchang Lee, Yeong Ouk Kim, Sang-Cheol Park and Jongsik Chun. OrthoANI: An improved algorithm and software for calculating average nucleotide identity (2016). International Journal of Systematic and Evolutionary Microbiology. doi:10.1099/ijsem.0.000760. PMID:26585518.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for orthoani-0.6.0-py2.py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 303db121e92a707154c99a1cea08217e7bc4499d59569eb30313427fdf06b96e |
|
| MD5 | b33f6a6338fca91b3ee0ab48e4901464 |
|
| BLAKE2b-256 | e3878c1ed0e7a9ad0500a11892a00b59df54877cc780116c88738b4908d4f67d |