'Library for producing, solving and visualising pharmacokinetic models.'
Project description
Pharmacokinetic Modelling Group Project
A PharmacoKinetic (PK) modelling function for analysis of injected solute dynamics over time, developed by Group 6 of the 2021 Software Engineering course. This model has been developed with the end-user in mind, and includes an easy-to-use interface to guide refinement of tissue models.
https://software-engineering-bbsrc-group-6.github.io/PK_modelling/
Authors
Amit Halkhoree - amit.halkhoree@dtc.ox.ac.uk
Cameron Anderson - cameron.anderson@dtc.ox.ac.uk
Dan Hudson - alexander.hudson@dtc.ox.ac.uk
Ishaan Kapoor - ishaan.kapoor@dtc.ox.ac.uk
Joseph Pollacco - joseph.pollacco@dtc.ox.ac.uk
Samuel Johnson - samuel.johnson@dtc.ox.ac.uk
Background
Ref: https://sabs-r3.github.io/software-engineering-projects/01-introduction/index.html.
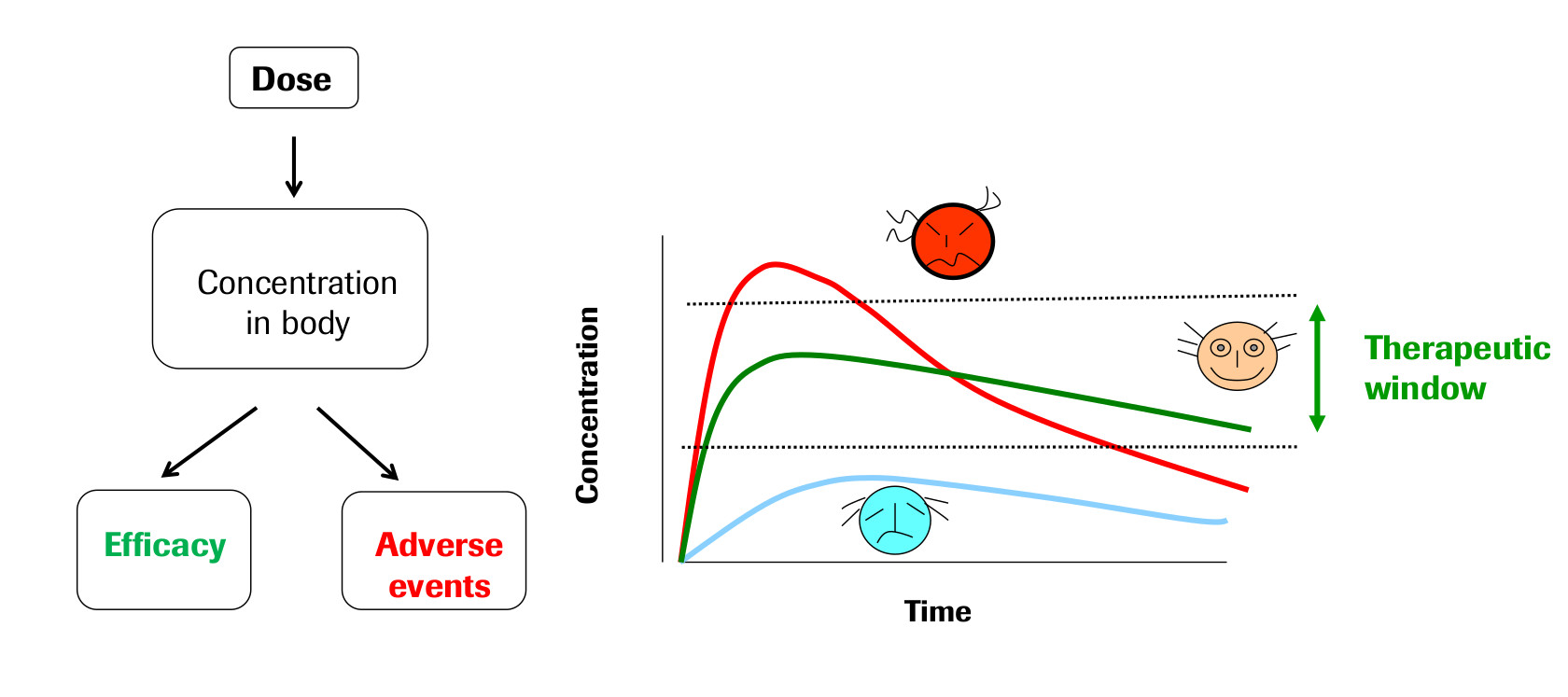
The field of Pharmacokinetics (PK) provides a quantitative basis for describing the delivery of a drug to a patient, the diffusion of that drug through the plasma/body tissue, and the subsequent clearance of the drug from the patient’s system. PK is used to ensure that there is sufficient concentration of the drug to maintain the required efficacy of the drug, while ensuring that the concentration levels remain below the toxic threshold. Pharmacokinetic (PK) models are often combined with Pharmacodynamic (PD) models, which model the positive effects of the drug, such as the binding of a drug to the biological target, and/or undesirable side effects, to form a full PKPD model of the drug-body interaction. This project will only focus on PK, neglecting the interaction with a PD model.
Model overview
Our model consists of three principal modules:
- A solver (solver.py) that computes the distribution of solute in different tissue compartments over time, given input parameters including the number of compartments to model, dose, mode of administration (intravenous or subcutaneous), compartment volume and transition rate constants
- A user interface (ux.py) that prompts the user to configure the model
- A visualisation module (visualiser.py) that generates and saves comparative plots to facilitate evaluation of PK using different administration methods
Folder structure
This repository includes the following key files/folders:
- .github/workflows: directory of github actions ensuring continuous integration (CI) of repository updates
- data: directory for saving outputs
- docs: files required for automated document production
- jsons: directory for saving user input and passing to the solver
- pk_model: directory for main executables
- definitons.py: Key helper functions for the solver
- dosing.py: computes dose changes for different administration models
- solver.py: Solver model for PK modelling
- ux.py: User interface executable
- visualiser.py Graph plotting function
- README.md: text file populating this guide
- tests: unit tests fed to pytest for continuous integration
- venv: virtual environment including dependencies for this package
- .gitignore: file controlling which files are/are not updated by git during development
- requirements.txt: Dependencies required for proper package functioning
Installation
Clone the git repository to your local directory:
'git clone https://github.com/Software-Engineering-BBSRC-Group-6/PK_modelling'
Set the correct working directory:
'cd PK_modelling'
We recommend then creating and activating a virtual environment
'python 3 venv venv'
'source venv/bin/activate'
The project dependencies can then be installed via pip
pip install oxbiodtp-7-PK-modelling
OR
'pip install -r requirements.txt' (this will also install modules required for development)'
Execute the main file to run the programme
python -m pk_model_main_pipeline.py \
Runtime
The model will then pose a series of questions, to be answered using the command line. An example runscript is provided below:
How many models would you like to test? 1
What kind of models do you want to build? (intravenous bolous (ib) / subcutaneous (sc)): ib
What compound or drug are you using? paracetamol
How is the dose delivered? Constantly over time (c), Instantaneously (i) or Repeated instantaneous doses (r): i
What is the mass of the dose of paracetamol that you want to test? (units in ng): 500
What time period would you like to simluate the model? (units in hours): 24
What interval time would you like in the simulation? (units in hours): 1
What is the clearance rate? (units in ng/hour): 50
Enter volume (L), transition rate (ng/hour) for the main compartment (all seperated by spaces - eg: 5 25 ): 7 50
How many peripheral compartments do you want to test?: 1
Enter volume (L), transition rate (ng/hour) of the compartment (all seperated by spaces - eg: 5 25): 2 25
Please enter the name of the compartment (please ensure correct spelling): heart
Would you like to generate a graph? (Y/N): y
This should produce a plot of the distribution of paracetamol between the main compartment and the heart, and save the resulting .csv in the data directory. NB: If running multiple models, please use the same number of compartments - developers are working to correct this issue.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Hashes for oxbiodtp_7_PK_modelling-1.0.0.tar.gz
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 109d2ba478d7c488afdd55cd3a5cab3a35eb6a23e461811f201def51420a35b0 |
|
| MD5 | b1aa62363ace3e4cebfe46892b275644 |
|
| BLAKE2b-256 | ac5852ceeeb0391325e8ab692c79277bbc618b88fdb241932d8c0b713a63aded |
Hashes for oxbiodtp_7_PK_modelling-1.0.0-py3-none-any.whl
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 0bdffbf4a82e1cf48f2e2c0832980498b1c9b251895a550f05d6c92753fa8b0b |
|
| MD5 | 0d364473eebe314a3ebf270f4d8c64e5 |
|
| BLAKE2b-256 | 578fb00a7d65c749b11387c30ea4a3d88655c3d665de4f513f8202a05d02f57b |