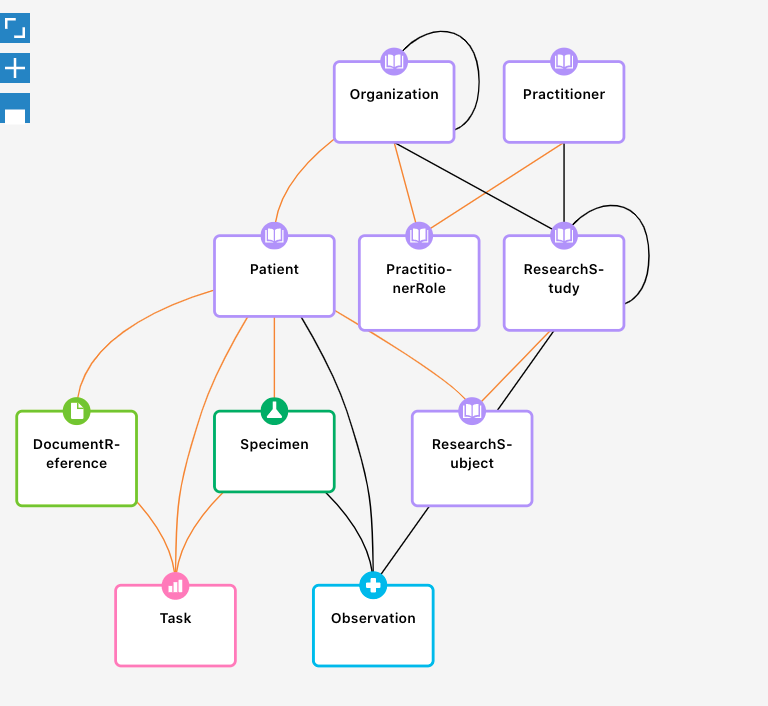
Render a PFB graph from FHIR data
Project description
Table of Contents
pfb_fhir
About The Project
Render a PFB graph from FHIR data
Built With
- pydantic
- fastavro
- pypfb[gen3]
- flatten_json
- fhirclientr4
- synthea
Special thanks
- Kyle Ellrott (OHSU)
- NHGRI Genomic Data Science Analysis, Visualization and Informatics Lab-space AnVIL
- NCPI FHIR Working Group, Robert Carroll (VUMC), Allison Heath (CHOP)
- Center for Translational Data Science (UChicago) gen3
- ncpi-fhir-pfb-prototype Brian O'Connor (Broad), Ann Van (Nimbus Informatics)
- GA4GH cohort-rep-hackathon synthea
Getting Started
Installation:
# clone
git clone https://github.com/bmeg/pfb_fhir
cd pfb_fhir
To get a local copy up and running follow these simple example steps.
# use virtual env
python3 -m venv venv
source venv/bin/activate
# install dependencies
pip install -r requirements.txt
# install this package
pip install -e .
Usage
See help for a list of commands, examine configuration examples in fixtures:
$ pfb_fhir --help
Usage: pfb_fhir [OPTIONS] COMMAND [ARGS]...
Render FHIR Data in PFB.
Options:
-l, --log-level [NOTSET|DEBUG|INFO|WARNING|ERROR|CRITICAL]
--output_path TEXT Output path for working files and output.
Read from PFB_FHIR_OUTPUT_PATH [default:
./DATA]
--config_path TEXT Path to config file. Read from
PFB_FHIR_CONFIG_PATH [default:
./config.yaml]
--help Show this message and exit.
Commands:
version Print the version.
transform Transform FHIR resources from directory.
inspect Inspect a PFB.
visualize Create a simple visualization.
config Print the config.
demo Download Test data and create example PFB and figure.
Environmental variables and their defaults
- PFB_FHIR_CONFIG_PATH
./config.yaml - PFB_FHIR_OUTPUT_PATH
DATA/ - PFB_FHIR_CACHE_PATH
cache/
Demos
Several demonstration datasets are available:
- anvil Read 1000G data from AnVIL.
- dbgap Read open access data from dbGAP's FHIR service.
- kf Read synthetic clinical data created by kids first.
- ncpi Read examples from ncpi ImplementationGuide.
- synthea Read synthetic clinical data created by synthea.
- genomic-reporting Read oncology example from ImplementationGuide.
For example:
# run the demo
pfb_fhir demo dbgap
# view the image
open DEMO/dbgap/output/dbgap.pfb.avro.png
# inspect the pfb
pfb_fhir inspect --pfb_path DEMO/dbgap/output/dbgap.pfb.avro
# use gen3's pfb utility
cat DEMO/dbgap/output/dbgap.pfb.avro | pfb show stats
# or native avro tools
java -jar avro-tools-1.11.0.jar getschema DEMO/dbgap/output/dbgap.pfb.avro | \
jq '.fields[] | select(.name == "object") | .type[] | .name '
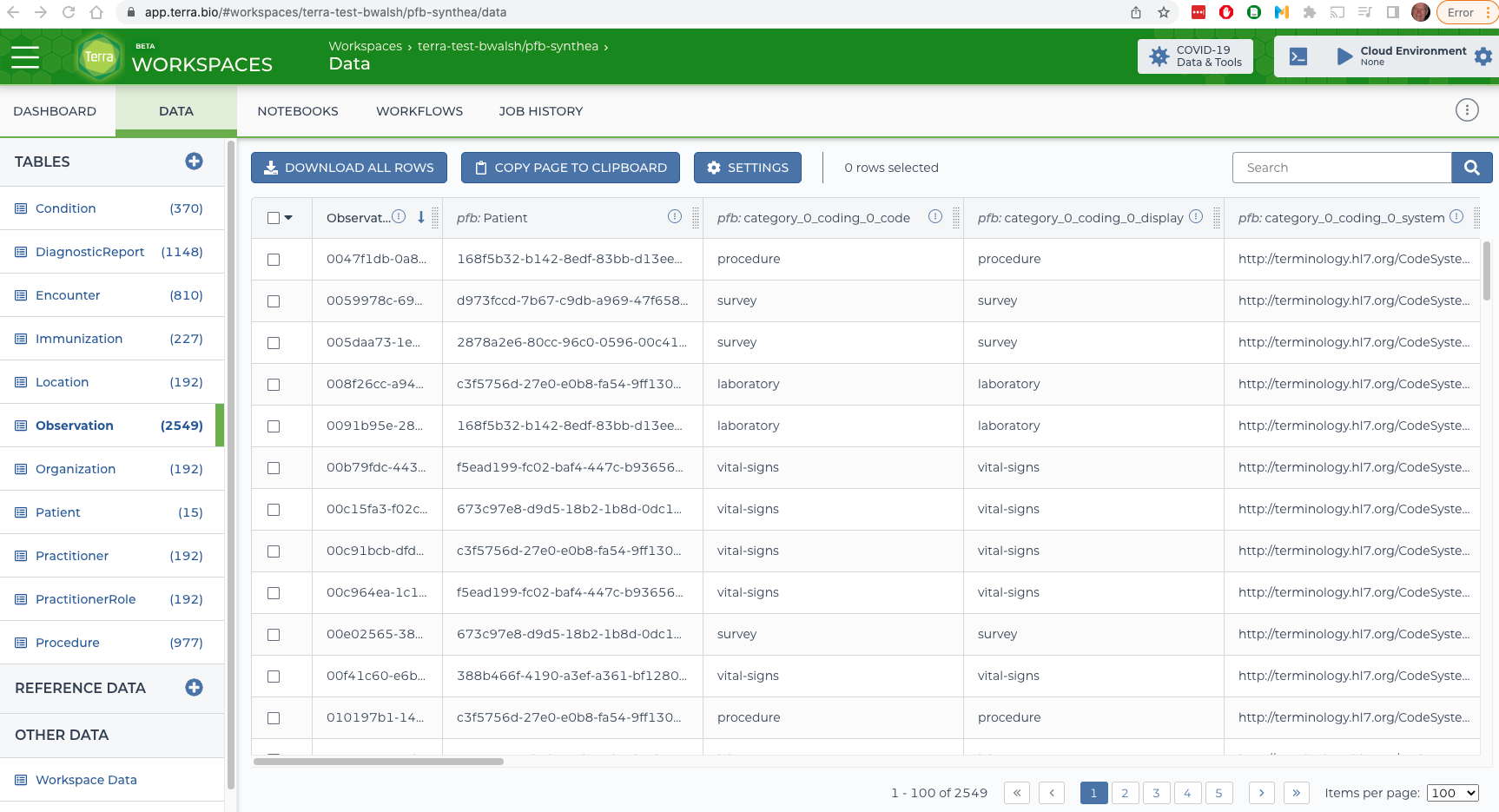
Using the PFB
Terra
# run the demo
pfb_fhir demo synthea
# upload that PFB to google storage
gsutil cp DEMO/synthea/output/synthea.pfb.avro $GOOGLE_BUCKET
# Sign the url so terra can read it.
gsutil signurl -u $GOOGLE_BUCKET/synthea.pfb.avro
# Or, grant access
# gsutil acl ch -u AllUsers:R $GOOGLE_BUCKET/synthea.pfb.avro
# url encode the signed url
open 'https://app.terra.bio/#import-data?format=PFB&url=https:....'
Gen3
The easiest way to see the resulting schema in gen3 is to use the Data Dictionary Development workflow Use the intermediate file located at <PFB_FHIR_OUTPUT_PATH>//output/dump-ordered.json e.g. DEMO/ncpi/output/dump-ordered.json
Contributing
Contributions are what make the open source community such an amazing place to learn, inspire, and create. Any contributions you make are greatly appreciated.
If you have a suggestion that would make this better, please fork the repo and create a pull request. You can also simply open an issue with the tag "enhancement". Don't forget to give the project a star! Thanks again!
- Fork the Project
- Create your Feature Branch (
git checkout -b feature/AmazingFeature) - Commit your Changes (
git commit -m 'Add some AmazingFeature') - Push to the Branch (
git push origin feature/AmazingFeature) - Open a Pull Request
Distribution
# update pypi
export TWINE_USERNAME= # the username to use for authentication to the repository.
export TWINE_PASSWORD= # the password to use for authentication to the repository.
rm -r dist/
python3 setup.py sdist bdist_wheel
twine upload dist/*
License
Distributed under the Apache License. See LICENSE for more information.
Contact
Brian Walsh - @bpwalsh - walsbr AT ohsu DOT edu
Acknowledgments
Roadmap
- ✅ config.yaml driven map of FHIR resource to PFB node,
- ✅ Parse FHIR resources, retrieve and cache FHIR profile (schema) elements,
- ✅ Recursively match with FHIR profiles (schemas)
- ✅ Flatten for export
- ✅ write to PFB
- ✅ test cases for protected/ resources - ResearchSubject, Patient, Specimen, Task, DocumentReference
- ✅ networkx based visualization
- ✅ pfb cli
- ✅ demo with public data
- ✅ update README with Terra import examples
- ✅ update README with gen3 import examples
- ✅ update to new release of pypfb
0.5.18 - ✅ include kids first
- ✅ simplify
- ✅ add genomic reporting examples
- ✅ pypi
Distribution
Test with docker
docker build -t pfb_fhir .
# typical run command
docker run -v $(pwd)/cache:/app/pfb_fhir/cache -v $(pwd)/DEMO:/app/pfb_fhir/DEMO pfb_fhir demo ncpi
Update pypi
export TWINE_USERNAME= # the username to use for authentication to the repository.
export TWINE_PASSWORD= # the password to use for authentication to the repository.
rm -r dist/
python3 setup.py sdist bdist_wheel
twine upload dist/*
Known Issues
simplify- could do a better job of making resulting nodes more "data frame friendly"config.yaml- currently there is no way of expressing a reference found in a FHIR extension.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.