A library for checking the limits of phylogenetic tree estimation.
Project description
phylim: a phylogenetic limit evaluation library built on cogent3
phylim evaluates the identifiability when estimating the phylogenetic tree using the Markov model. The identifiability is the key condition of the Markov model used in phylogenetics to fulfil consistency.
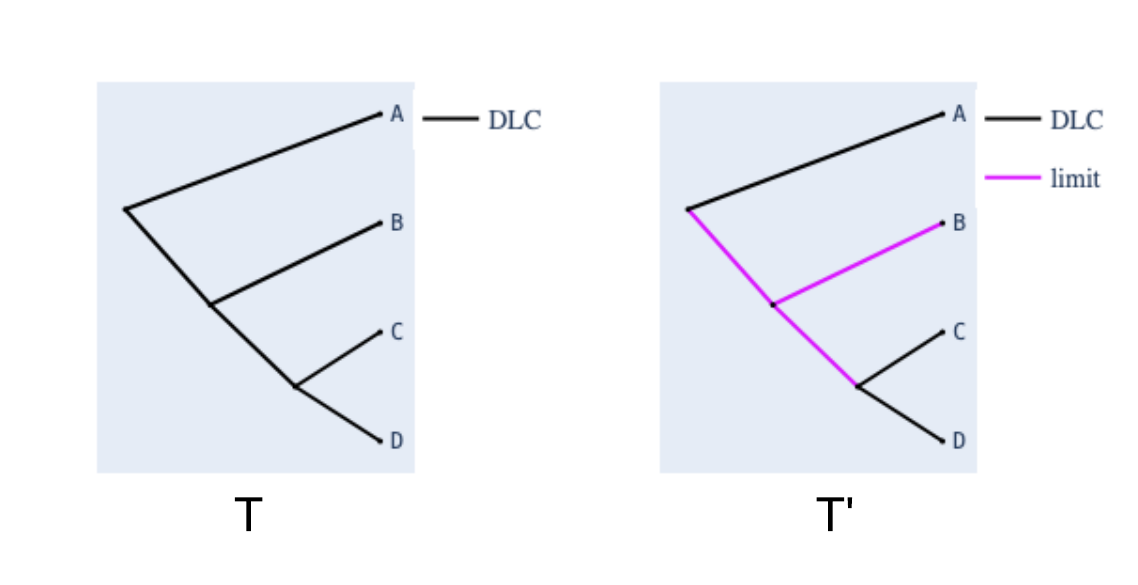
Establishing identifiability relies on the arrangement of specific types of transition probability matrices (e.g., DLC and sympathetic) while avoiding other types. A key concern arises when a tree does not meet the condition that, for each node, a path to a tip must exist where all matrices along the path are DLC. Such trees are not identifiable 🪚🎄! For instance, in the figure below, tree T' contains a node surrounded by a specific type of non-DLC matrix, rendering it non-identifiable. In contrast, compare T' with tree T.
phylim provides a quick, handy method to check the identifiability of a model fit, where we developed a main cogent3 app, phylim. phylim is compatible with piqtree, a python library that exposes features from iqtree2.
The following content will demonstrate how to set up phylim and give some tutorials on the main identifiability check app and other associated apps.
Installation
pip install phylim
Let's see if it has been done successfully. In the package directory:
pytest
Hope all tests passed! :white_check_mark: :blush:
Run the check of identifiability
If you fit a model to an alignment and get the model result:
>>> from cogent3 import get_app, make_aligned_seqs
>>> aln = make_aligned_seqs(
... {
... "Human": "ATGCGGCTCGCGGAGGCCGCGCTCGCGGAG",
... "Gorilla": "ATGCGGCGCGCGGAGGCCGCGCTCGCGGAG",
... "Mouse": "ATGCCCGGCGCCAAGGCAGCGCTGGCGGAG",
... },
... info={"moltype": "dna", "source": "foo"},
... )
>>> app_fit = get_app("model", "GTR")
>>> result = app_fit(aln)
You can easily check the identifiability by:
>>> checker = get_app("phylim")
>>> checked = checker(result)
>>> checked.is_identifiable
True
The phylim app wraps all information about phylogenetic limits.
>>> checked
| Source | Model Name | Identifiable | Has Boundary Values | Version |
|---|---|---|---|---|
| brca1.fasta | GTR | True | True | 2025.1.12 |
You can also use features like classifying all matrices or checking boundary values in a model fit.
Label all transition probability matrices in a model fit
You can call classify_model_psubs to give the category of all the matrices:
>>> from phylim import classify_model_psubs
>>> labelled = classify_model_psubs(result)
>>> labelled
| edge name | matrix category |
|---|---|
| Gorilla | DLC |
| Human | DLC |
| Mouse | DLC |
Check if all parameter fits are within the boundary
>>> from phylim import check_fit_boundary
>>> violations = check_fit_boundary(result)
>>> violations
BoundsViolation(source='foo', vio=[{'par_name': 'C/T', 'init': np.float64(1.0000000147345554e-06), 'lower': 1e-06, 'upper': 50}, {'par_name': 'A/T', 'init': np.float64(1.0000000625906854e-06), 'lower': 1e-06, 'upper': 50}])
❗For users who want to check identifiability on a model with multiple likelihood functions (e.g. split codon model), please check https://github.com/HuttleyLab/PhyLim/issues/23#issuecomment-3125670158
Check identifiability for piqtree
phylim provides an app, phylim_to_model_result, which allows you to build the likelihood function from a piqtree output tree.
>>> phylo = get_app("piq_build_tree", model="GTR")
>>> tree = phylo(aln)
>>> lf_from = get_app("phylim_to_model_result")
>>> result = lf_from(tree)
>>> checker = get_app("phylim")
>>> checked = checker(result)
>>> checked.is_identifiable
True
Colour the edges for a phylogenetic tree based on matrix categories
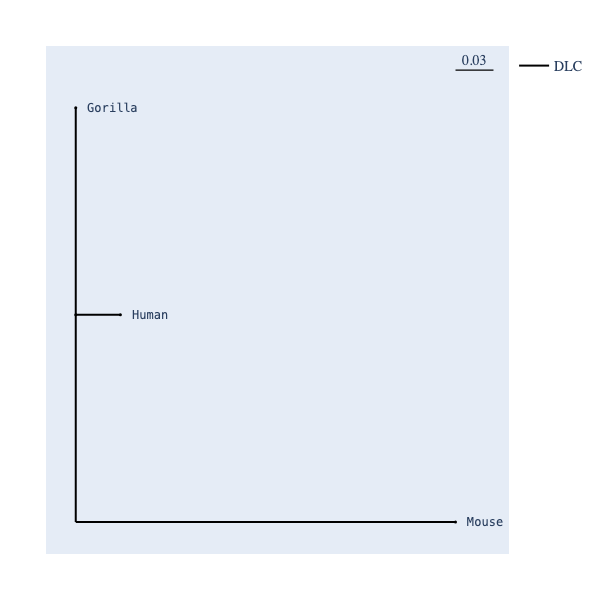
If you obtain a model fit, phylim can visualise the tree with labelled matrices.
phylim provides an app, phylim_style_tree, which takes an edge-matrix category map and colours the edges:
>>> from phylim import classify_model_psubs
>>> edge_to_cat = classify_model_psubs(result)
>>> tree = result.tree
>>> tree_styler = get_app("phylim_style_tree", edge_to_cat)
>>> tree_styler(tree)
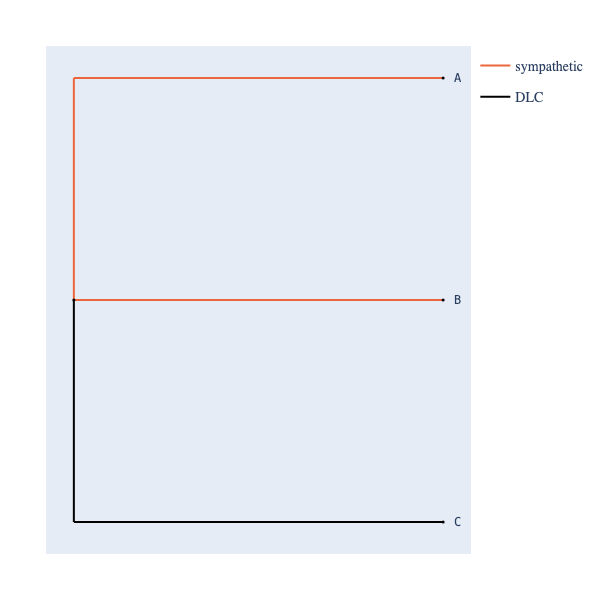
You can also colour edges using a user-defined edge-matrix category map, applicable to any tree object!
>>> from cogent3 import make_tree
>>> from phylim import SYMPATHETIC, DLC
>>> tree = make_tree("(A, B, C);")
>>> edge_to_cat = {"A":SYMPATHETIC, "B":SYMPATHETIC, "C":DLC}
>>> tree_styler = get_app("phylim_style_tree", edge_to_cat)
>>> tree_styler(tree)
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file phylim-2025.8.27.tar.gz.
File metadata
- Download URL: phylim-2025.8.27.tar.gz
- Upload date:
- Size: 12.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.12.9
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
e6a9d19aa8c018f397aeb82e1a7c99699248c496c61e81c78846e575388e76ae
|
|
| MD5 |
811aa04ebf5b1ea2a4b5169acd14b75d
|
|
| BLAKE2b-256 |
0b9470afe7a3ee9e3e2720edfa4cfd2b1ebc1832d77da4c0f68c491f089c6d31
|
Provenance
The following attestation bundles were made for phylim-2025.8.27.tar.gz:
Publisher:
release.yml on HuttleyLab/PhyLim
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
phylim-2025.8.27.tar.gz -
Subject digest:
e6a9d19aa8c018f397aeb82e1a7c99699248c496c61e81c78846e575388e76ae - Sigstore transparency entry: 437991368
- Sigstore integration time:
-
Permalink:
HuttleyLab/PhyLim@18c7375ea4f582c30c72b82343c2c954dfd8b3bc -
Branch / Tag:
refs/tags/2025.8.27 - Owner: https://github.com/HuttleyLab
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
release.yml@18c7375ea4f582c30c72b82343c2c954dfd8b3bc -
Trigger Event:
workflow_dispatch
-
Statement type:
File details
Details for the file phylim-2025.8.27-py3-none-any.whl.
File metadata
- Download URL: phylim-2025.8.27-py3-none-any.whl
- Upload date:
- Size: 12.2 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/6.1.0 CPython/3.12.9
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
5b26d14f66b0221bdec0e45095a7bd75b02d1dc5e46a1d9cdc6ef189eef56ca3
|
|
| MD5 |
3d7ff11874a2f84dd3c5ad04364dfa45
|
|
| BLAKE2b-256 |
b0181d56ae9dca2a3a588bde7c208515423cfb646c892f79e17e247cee286cb9
|
Provenance
The following attestation bundles were made for phylim-2025.8.27-py3-none-any.whl:
Publisher:
release.yml on HuttleyLab/PhyLim
-
Statement:
-
Statement type:
https://in-toto.io/Statement/v1 -
Predicate type:
https://docs.pypi.org/attestations/publish/v1 -
Subject name:
phylim-2025.8.27-py3-none-any.whl -
Subject digest:
5b26d14f66b0221bdec0e45095a7bd75b02d1dc5e46a1d9cdc6ef189eef56ca3 - Sigstore transparency entry: 437991374
- Sigstore integration time:
-
Permalink:
HuttleyLab/PhyLim@18c7375ea4f582c30c72b82343c2c954dfd8b3bc -
Branch / Tag:
refs/tags/2025.8.27 - Owner: https://github.com/HuttleyLab
-
Access:
public
-
Token Issuer:
https://token.actions.githubusercontent.com -
Runner Environment:
github-hosted -
Publication workflow:
release.yml@18c7375ea4f582c30c72b82343c2c954dfd8b3bc -
Trigger Event:
workflow_dispatch
-
Statement type:















