No project description provided
Project description


Pyinterpolate
version 1.2.1
Important notice
The package was updated to version 1.0 in June 2025. There are breaking API changes, so please, refer to the CHANGELOG to know more about the changes.
Citation
Moliński, S., (2022). Pyinterpolate: Spatial interpolation in Python for point measurements and aggregated datasets. Journal of Open Source Software, 7(70), 2869, https://doi.org/10.21105/joss.02869
Bibtex
@article{Moliński2022,
doi = {10.21105/joss.02869},
url = {https://doi.org/10.21105/joss.02869},
year = {2022},
publisher = {The Open Journal},
volume = {7},
number = {70},
pages = {2869},
author = {Moliński, Szymon},
title = {Pyinterpolate: Spatial interpolation in Python for point measurements and aggregated datasets},
journal = {Journal of Open Source Software} }
Introduction
Pyinterpolate is the Python library for spatial statistics. The package provides access to spatial statistics tools (variogram analysis, Kriging, Poisson Kriging, Indicator Kriging, Inverse Distance Weighting).
If you’re:
- GIS expert
- Geologist
- Social scientist
Then you might find this package useful. The core functionalities of Pyinterpolate are spatial interpolation and spatial prediction for point and block datasets.
Pyinterpolate performs:
- Ordinary Kriging and Simple Kriging - spatial interpolation from points
- Centroid-based Poisson Kriging of polygons - spatial interpolation from blocks and regions
- Area-to-area and Area-to-point Poisson Kriging of Polygons - spatial interpolation and data deconvolution from areas to points
- Indicator Kriging - kriging based on probabilities
- Universal Kriging - kriging with trend
- Inverse Distance Weighting - benchmarking spatial interpolation technique
- Semivariogram regularization and deconvolution - transforming variogram of areal data in regards to point support data
- Semivariogram modeling and analysis - is your data spatially correlated? How do neighbors influence each other?
How does it work?
The package has multiple spatial interpolation functions. The flow of analysis is usually the same for each method:
[1.] Load your dataset with GeoPandas or numpy.
import geopandas as gpd
point_data = gpd.read_file('dem.gpkg') # x (lon), y (lat), value
[2.] Pass loaded data to pyinterpolate, calculate experimental variogram.
from pyinterpolate import ExperimentalVariogram
step_size = 500
max_range = 40000
experimental_variogram = ExperimentalVariogram(
ds=point_data,
step_size=step_size,
max_range=max_range
)
[3.] Fit experimental semivariogram to theoretical model, it is equivalent of the fit() method known from machine learning packages.
from pyinterpolate import build_theoretical_variogram
sill = experimental_variogram.variance
nugget = 0
variogram_range = 8000
semivar = build_theoretical_variogram(
experimental_variogram=experimental_variogram,
models_group='linear',
nugget=nugget,
rang=variogram_range,
sill=sill
)
[4.] Interpolate values in unknown locations.
from pyinterpolate import ordinary_kriging
unknown_point = (20000, 65000)
prediction = ordinary_kriging(theoretical_model=semivar,
known_locations=point_data,
unknown_location=unknown_point,
no_neighbors=32)
[5.] Analyze error and uncertainty of predictions.
print(prediction) # [predicted, variance error, lon, lat]
>> [211.23, 0.89, 20000, 60000]
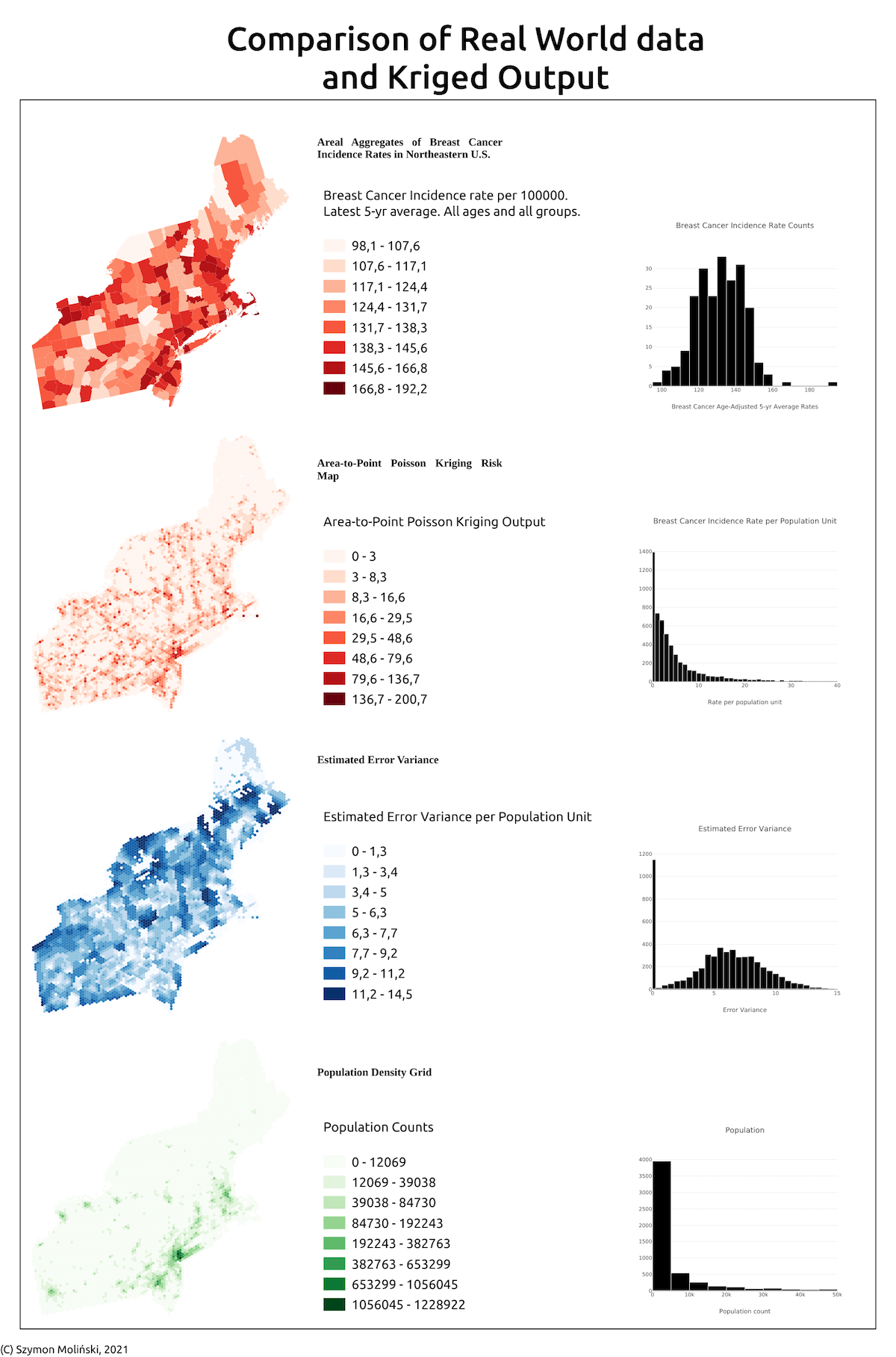
With Pyinterpolate you can analyze and transform aggregated data. Here is the example of spatial disaggregation of areal data into point support using Poisson Kriging:
Status
Operational: no API changes in the current release cycle.
Setup
Setup with conda: conda install -c conda-forge pyinterpolate
Setup with pip: pip install pyinterpolate
Detailed instructions on how to install the package are presented in the file SETUP.md. We pointed out there most common problems related to third-party packages.
You may follow those setup steps to create a conda environment with the package for your work:
Recommended - conda installation
[1.] Create conda environment with Python >= 3.10
conda create -n [YOUR ENV NAME] -c conda-forge python=3.10 pyinterpolate
[2.] Activate environment.
conda activate [YOUR ENV NAME]
[3.] You are ready to use the package!
pip installation
With Python>=3.9 and system libspatialindex_c.so dependencies you may install package by simple command:
pip install pyinterpolate
A world of advice, you should always use Virtual Environment for the installation. You may consider using PipEnv too.
Tests and contribution
All tests are grouped in the test directory. If you would like to contribute, then you won't avoid testing, but it is described step-by-step here: CONTRIBUTING.md
Use cases
- Lyme Disease Risk Assessment Based on the Earth Observation Data and Spatial Processing Technique. ESA Φ-week 2021
- Disaggregation of socio-economic data
- Interpolation of air pollution
- Educational - Spatial Data Science for Social Geography Course - Interpolation
Bibliography
Pyinterpolate was created thanks to many resources and all of them are pointed here:
- Armstrong M., Basic Linear Geostatistics, Springer 1998,
- GIS Algorithms by Ningchuan Xiao: https://uk.sagepub.com/en-gb/eur/gis-algorithms/book241284
- Pardo-Iguzquiza E., VARFIT: a fortran-77 program for fitting variogram models by weighted least squares, Computers & Geosciences 25, 251-261, 1999,
- Goovaerts P., Kriging and Semivariogram Deconvolution in the Presence of Irregular Geographical Units, Mathematical Geology 40(1), 101-128, 2008
- Deutsch C.V., Correcting for Negative Weights in Ordinary Kriging, Computers & Geosciences Vol.22, No.7, pp. 765-773, 1996
Community
Join our community in Discord: Discord Server Pyinterpolate
Requirements and dependencies (v 1.x)
Core requirements and dependencies are:
- Python >= 3.10
- geopandas
- matplotlib
- numpy
- prettytable
- pydantic
- scipy
- tqdm
You may check a specific version of requirements in the setup.cfg file. Required packages versions are updated in a regular interval.
Package structure
High level overview:
-
pyinterpolate-
core- data structures and models, data processing pipelines -
distance- distance and angles -
evaluate- cross-validation and modeling metrics -
idw- inverse distance weighting -
kriging- Ordinary Kriging, Simple Kriging, Poisson Kriging: centroid based, area-to-area, area-to-point, Indicator Kriging -
transform- internal data processing functions -
semivariogram- experimental variogram, theoretical variogram, variogram point cloud, semivariogram regularization & deconvolution, indicator variogram -
viz- interpolation of smooth surfaces from points into rasters.
-
-
tutorials-
api-examples- tutorials covering the API -
functional- tutorials covering concrete use cases
-
Datasets
Datasets and scripts to download spatial data from external API's are available in a dedicated package: pyinterpolate-datasets
API documentation
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file pyinterpolate-1.2.1.tar.gz.
File metadata
- Download URL: pyinterpolate-1.2.1.tar.gz
- Upload date:
- Size: 129.4 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.0.1 CPython/3.12.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
5ac5a1c778deda27bc740b33cbe746472c8e655bb90e52934d14a283779a99f6
|
|
| MD5 |
399a8b0cf171bd6d2355eaedbb38ba6d
|
|
| BLAKE2b-256 |
19bf81b14fa105c4acf303bf97958cc03c97b2ec0912af5219307679894ad81f
|
File details
Details for the file pyinterpolate-1.2.1-py3-none-any.whl.
File metadata
- Download URL: pyinterpolate-1.2.1-py3-none-any.whl
- Upload date:
- Size: 170.8 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.0.1 CPython/3.12.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
b8130c1062fa3feb1d64e1e577febddddaf27140b35cf1f424a5fb301d9e86aa
|
|
| MD5 |
8dbe1f7fb4f9603f9a4a5b65a6b79c2d
|
|
| BLAKE2b-256 |
6196e621e0653a13db79a75eab1f9b5e5901b97402a77f09cce8cccee0a1163c
|