A python library for nuclear astrophysics
Project description
pynucastro
pynucastro is a python library for nuclear astrophysics. It provides access to nuclear data and reaction rates, and tools for building and interactively exploring nuclear reaction networks.
The main features are:
-
Easy access to nuclear properties, including T-dependent partition functions, spins, masses, etc.
-
The ability to create a reaction network based on a collection of rates, a set of nuclei, or an arbitrary filter applied to a library.
- Includes support for ReacLib rates and tabulated weak rates from a variety of sources.
-
Interactive exploration of rates and networks in Jupyter notebooks, including:
-
Many different ways of visualizing a network.
-
The ability to export a NetworkX graph (for instance, to find cycles).
-
-
Methods to determine which rates are important and which might be missing from a network, as well as analyze the stiffness of a network.
-
The ability to approximate rates and derive reverse rates via detailed balance.
-
An NSE solver to find the equilibrium abundance of a set of nuclei given a thermodynamic state.
-
The ability to write out python or C++ code needed to integrate the network.
-
Additional physics methods for thermal neutrino sources and Fermi-Dirac integrals.
Documentation
Documentation for pynucastro is available here:
https://pynucastro.github.io/pynucastro/
Examples
Exploring a reaction rate
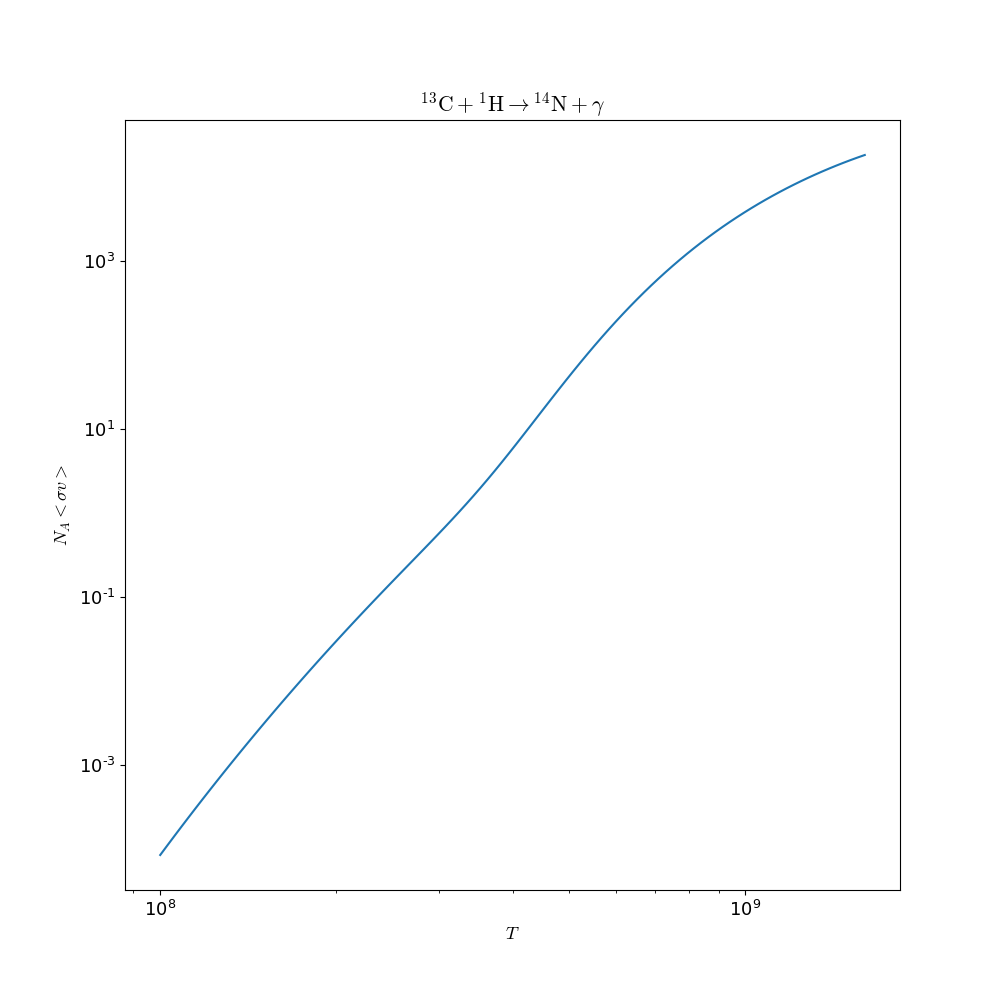
The following example reads in the ReacLib rate database and gets the rate for C13(p,g)N14 and evaluates it at a temperature of 1.e9 K and makes a plot of the T dependence:
In [1]: import pynucastro as pyna
In [2]: rl = pyna.ReacLibLibrary()
In [3]: c13pg = rl.get_rate_by_name("c13(p,g)n14")
In [4]: c13pg.eval(1.e9)
Out[4]: 3883.4778216250666
In [5]: fig = c13pg.plot()
In [6]: fig.savefig("c13pg.png")
The resulting figure is:
Creating a network
The following builds a simple network that has carbon burning, outputs the value of the rates at a set of thermodynamic conditions, and plots the network:
In [1]: import pynucastro as pyna
In [2]: nuclei = ["p", "he4", "c12", "n13", "o16", "ne20", "na23", "mg24"]
In [3]: net = pyna.network_helper(nuclei, use_tabular_rates=False)
In [4]: rho = 1.e7
In [5]: T = 3.e9
In [6]: comp = pyna.Composition(nuclei)
In [7]: comp.set_equal()
In [8]: net.evaluate_rates(rho=rho, T=T, composition=comp)
Out[8]:
{C12 + p ⟶ N13 + 𝛾: 52860361.23712939,
C12 + He4 ⟶ O16 + 𝛾: 231.46132999413808,
O16 + He4 ⟶ Ne20 + 𝛾: 4660.126437920582,
Ne20 + He4 ⟶ Mg24 + 𝛾: 53650.76987097864,
Na23 + p ⟶ Mg24 + 𝛾: 71891152.99067408,
C12 + C12 ⟶ p + Na23: 209.07753161235584,
C12 + C12 ⟶ He4 + Ne20: 259.65215046037304,
N13 + He4 ⟶ p + O16: 355698292.6571981,
O16 + C12 ⟶ He4 + Mg24: 0.8082923986833515,
Na23 + p ⟶ He4 + Ne20: 6840366554.79809,
3 He4 ⟶ C12 + 𝛾: 0.12389170353139083,
N13 ⟶ p + C12: 984313528.3619095,
O16 ⟶ He4 + C12: 0.00013799339351820605,
Ne20 ⟶ He4 + O16: 39.38899256329942,
Mg24 ⟶ p + Na23: 6.121492897494776e-07,
Mg24 ⟶ He4 + Ne20: 9.835129431742516e-06,
C12 ⟶ 3 He4: 0.008264918834094248,
O16 + p ⟶ He4 + N13: 11.428884290229457,
Ne20 + He4 ⟶ p + Na23: 317753.8001151714,
Ne20 + He4 ⟶ C12 + C12: 1.929507593099608e-05,
Na23 + p ⟶ C12 + C12: 0.33819870621531356,
Mg24 + He4 ⟶ C12 + O16: 1.8802924022864885e-11}
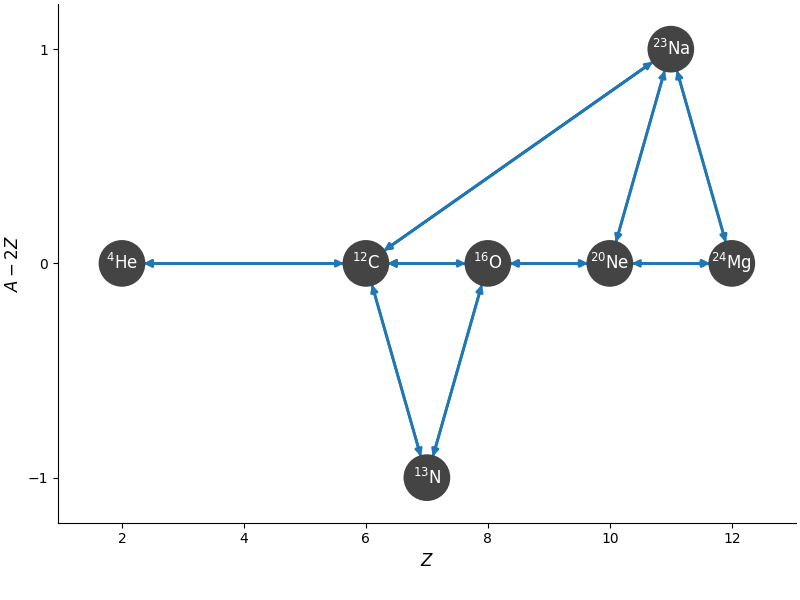
In [9]: fig = net.plot(rotated=True, hide_xalpha=True)
In [10]: fig.savefig("c-net.png")
Here network_helper found all of the rates that link the input
nuclei and rederived the reverse rates to be in detailed balance with
the forward rates.
The resulting figure is:
Interactive exploration
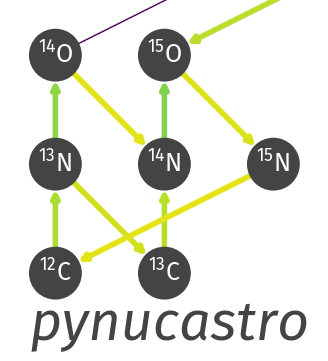
We can also interactively explore a reaction network. Here's an example of hot-CNO with breakout reactions:
by increasing the temperature, you can see the transition from CNO to hot-CNO (proton captures on C13 become faster than the beta decay) and then the breakout of hot-CNO when the alpha capture on O14 becomes faster than its decay.
Installation
pynucastro is available on PyPI and can be installed via:
pip install pynucastro
Alternately, to install via source, you can do:
pip install .
for a systemwide install, or
pip install --user .
for a single-user install. This will put the pynucastro modules and library in the default location python searches for packages.
Requirements
pynucastro is supported on Python 3.10 or later and the following libraries:
-
numpy -
sympy -
scipy -
matplotlib -
networkx -
ipywidgets -
numba
To build the documentation or run the unit tests, sphinx and
pytest are additionally required along with some supporting
packages. See the included requirements.txt file for a list of these
packages and versions. To install the packages from the requirements
file, do:
pip install -r requirements.txt
Is important to stress out that all the dependencies should be
installed before pynucastro, otherwise the installation should be
repeated.
Contributing
We welcome contributions from anyone, including posting issues or submitting pull requests to the pynucastro github. For more details on how to contribute to pynucastro, please see CONTRIBUTING.md.
Core Developers
People who make a number of substantive contributions (new features, bug fixes, etc.) will be named "core developers" of pynucastro.
Core developers will be recognized in the following ways:
-
invited to the group's slack team
-
listed in the User's Guide and website as a core developer
-
listed in the author list on the Zenodo DOI for the project (as given in the .zenodo.json file)
-
invited to co-author general code papers / proceedings describing pynucastro. (Note: science papers that use pynucastro will always be left to the science paper lead author to determine authorship).
If a core developer is inactive for 3 years, we may reassess their status as a core developer.
Getting Help
We use our Github Discussions page for requesting help and interacting with the community.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file pynucastro-2.9.0.tar.gz.
File metadata
- Download URL: pynucastro-2.9.0.tar.gz
- Upload date:
- Size: 50.3 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.14.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
089a306e6e9adc781f46426648c8258201003d31556a185cf247e6095ab6464f
|
|
| MD5 |
e152fd0cb7ed148c8a5bd032adbefb87
|
|
| BLAKE2b-256 |
cb51e8a3ee7fd9a8865e0f5a5d30d6edda12245dc126130cd826efc7b935d462
|
File details
Details for the file pynucastro-2.9.0-py3-none-any.whl.
File metadata
- Download URL: pynucastro-2.9.0-py3-none-any.whl
- Upload date:
- Size: 44.4 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/6.2.0 CPython/3.14.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
9cc8c2ac4d7b1e9dc382426f3f23037a681fb69f4679f5462dcfd87bcc373edc
|
|
| MD5 |
4afd6b4db107dfa3a924212364cac3e8
|
|
| BLAKE2b-256 |
d55f3828d8acae5df255cdc36bd16a89a4861f57e9bf812d768902239a731645
|