Ramachandran plotting tool
Project description
Ramachandran plotting tool
Draws a Ramachandran plot based on the input PDB file (e.g. 1mbn.pdb). Makes use of a Gaussian KDE (kernel density
estimation) to plot the density of favoured torsion angles (φ and ψ).
| Single mode | Batch mode |
|---|---|
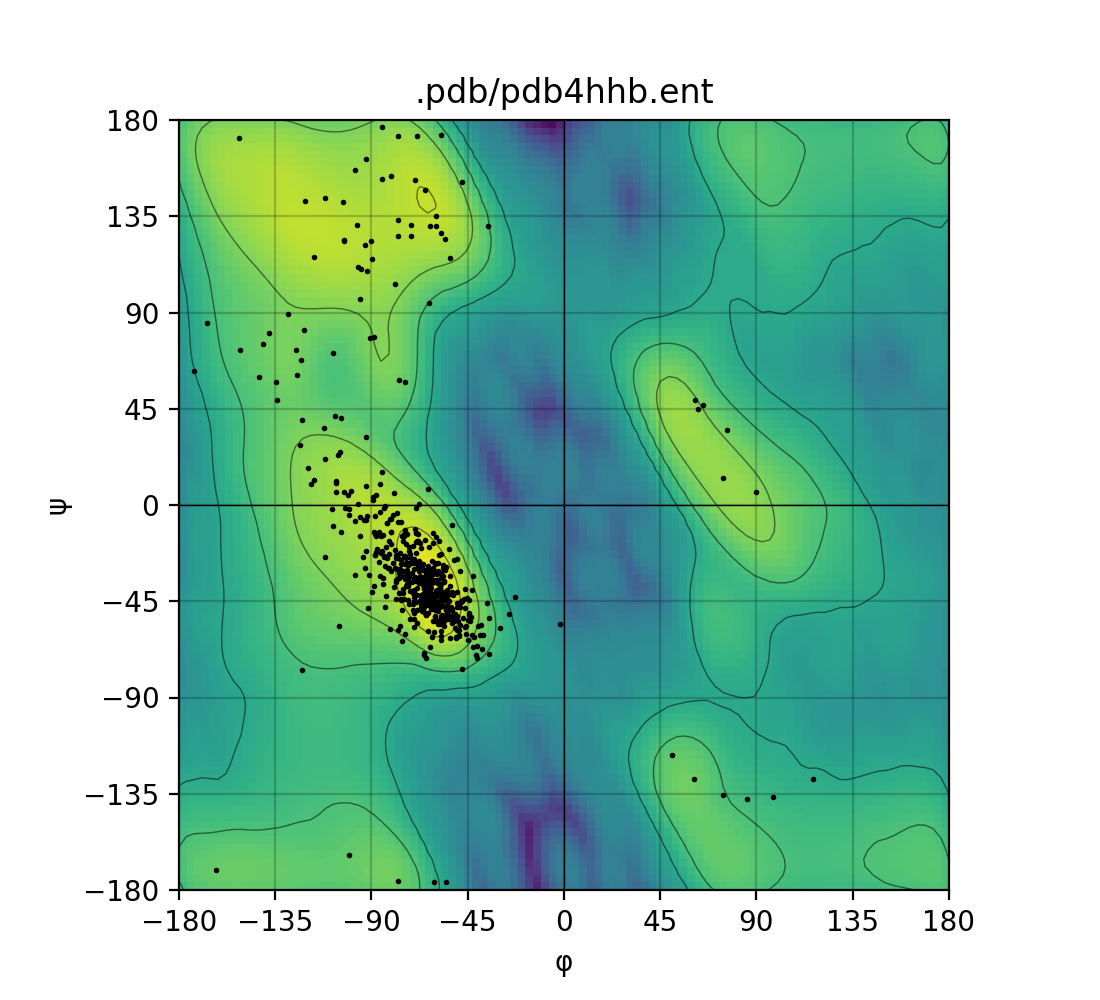 |
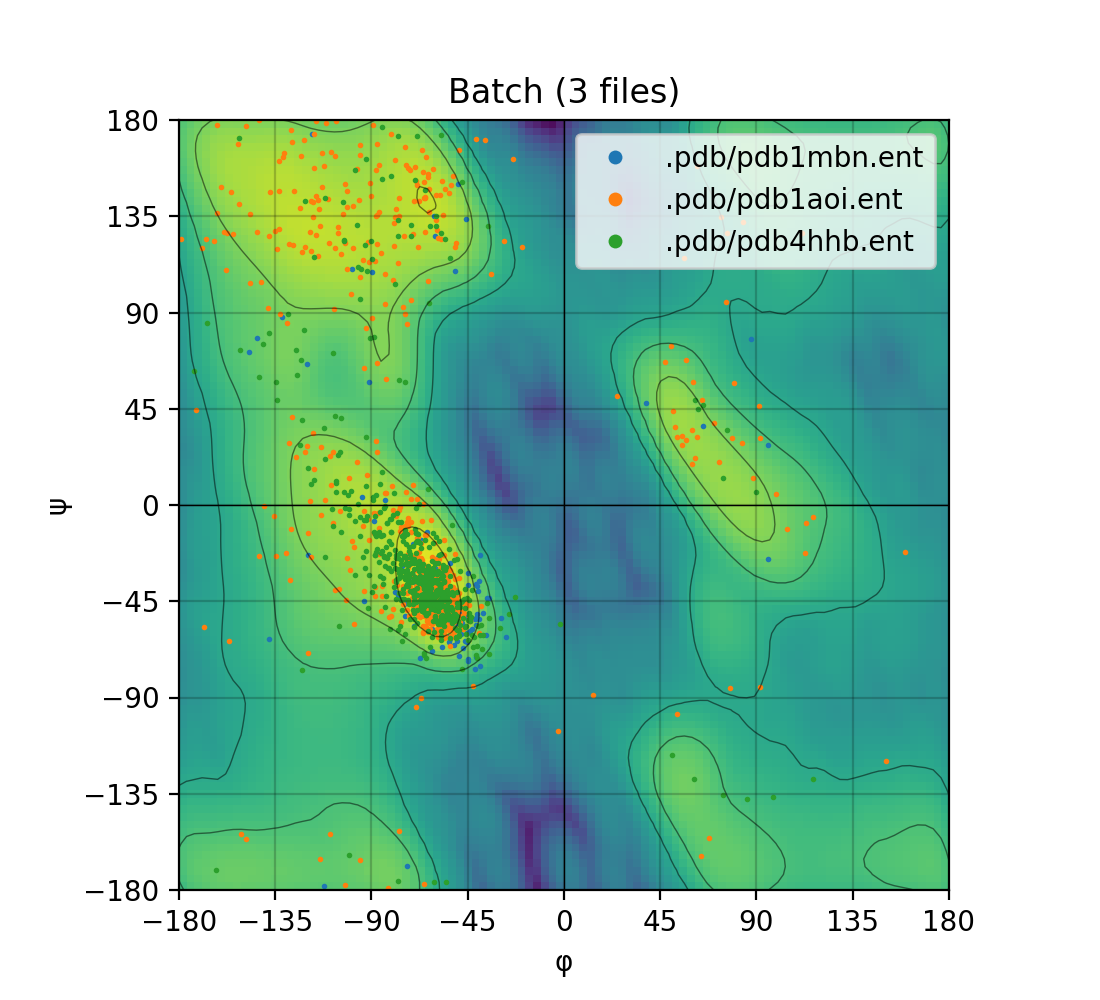 |
Installation
RamachanDraw is hosted on PyPi.
pip install ramachandraw
Usage
RamachanDraw includes useful functions to effortlessly draw a Ramachandran plot.
Fetch the PDB file
To draw a Ramachandran plot, we need a PDB file. RamachanDraw conveniently includes a function to automatically fetch and locally store the PDB file for the given PDB id.
Example
from ramachandraw.utils import fetch_pdb
fetch_pdb(pdb_id, verbose)
pdb_id (str|list|tuple): PDB id(s) corresponding to the PDB file(s) to be downloadedverbose (bool)(optional): set the verbosity, defaults toFalse- Returns: path(s) to PDB file(s)
Extract φ and ψ torsion angles
RamachanDraw extracts the φ and ψ torsion angles from the PDB file by taking advantage of the Biopython module. Additionally, aminoacid residues that were not drawn on the plot can be extract using the return_ignored argument.
Example
from ramachandraw.parser import get_phi_psi
phi_psi(pdb_filepath, prune, hide_warnings)
pdb_id (str|list|tuple): PDB filepath(s)prune (bool)(optional): prunes aminoacids with missing torsion angle(s), defaults toTruehide_warnings (bool)(optional): sets the verbosity of the PDB structure parser, defaults toTrue- Returns: Dictionary with keys as aminoacid residues and values as (φ, ψ) angles.
Ramachandran plot
Makes use of the matplotlib module to draw a highly customizable Ramachandran plot.
Example
from ramachandraw.utils import plot
plot(pdb_filepath, cmap="viridis", alpha=0.75, dpi=100, save=True, show=False, filename="plot.png")
pdb_file (str|list|tuple): PDB id(s) corresponding to the PDB entry to be downloadedcmap (str)(optional): colormap to be used (frommatplotlib) for the density of the favoured ("allowed") regions; default is viridis.alpha (float)(optional): sets the opacity of the colormap (value between 0-1); default is 0.75.dpi (int)(optional): resolution (in dots per inch); default is100.save (bool)(optional):True: saves the plot locally; default is True.
show (bool)(optional):True: shows the plot using the Qt5Agg backend; default is False.
filename (str)(optional): filename to be used in case the plot is saved (i.e.save=True); default isplot.png.- Returns: Ramachandran plot (
matplotlib.axes.Axesobject) that can be further customized if needed
Example
Herein you will find an example from the PDB id corresponding to the myoglobin entry: 1MBN - in the Protein Data Bank.
Single PDB
from ramachandraw.parser import get_phi_psi
from ramachandraw.utils import fetch_pdb, plot
# PDB id
pdb_id = "1mbn"
# Draw the Ramachandran plot
plot(fetch_pdb(pdb_id))
# Generate a dictionary to store the (phi, psi) torsion angles
torsion_angles = get_phi_psi(fetch_pdb(pdb_id))
Batch of PDBs
from ramachandraw.parser import get_phi_psi
from ramachandraw.utils import fetch_pdb, plot
# PDB id
pdb_ids = ["1mbn", "4hhb"]
# Draw the Ramachandran plot
plot(fetch_pdb(pdb_ids))
# Generate a list of dictionaries to store the (phi, psi) torsion angles
torsion_angles = get_phi_psi(fetch_pdb(pdb_ids))
Contributing
Feedback and constructive criticism is welcome. If necessary, open an issue in the issues tab.
License
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file ramachandraw-1.1.0.tar.gz.
File metadata
- Download URL: ramachandraw-1.1.0.tar.gz
- Upload date:
- Size: 84.9 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/2.3.2 CPython/3.14.3 Darwin/25.2.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
23b82ec8fe3071ad2d542ef8f9cd6c66a2354afdd5de0124035e46447ce34bd7
|
|
| MD5 |
323dcb963a99175f8230cb37ac833227
|
|
| BLAKE2b-256 |
fb6df94f5b5a46d255e82344541b3348f156cf94d9b30f084a760a4bdcf62df1
|
File details
Details for the file ramachandraw-1.1.0-py3-none-any.whl.
File metadata
- Download URL: ramachandraw-1.1.0-py3-none-any.whl
- Upload date:
- Size: 83.9 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/2.3.2 CPython/3.14.3 Darwin/25.2.0
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
01357b98f836065cc34199a5cbadf0968d3c68e95314d8a03fa0df07037b2c4f
|
|
| MD5 |
277142ef8a057132f17eb62e96f61f68
|
|
| BLAKE2b-256 |
60056425a94966e393fd128c2285f7b40cf0c7a3725b4ace13f4adb89d94068d
|



















