A very simple tool that compresses the overall size of the ONNX model by aggregating duplicate constant values as much as possible. Simple Constant value Shrink for ONNX.
Project description
scs4onnx
A very simple tool that compresses the overall size of the ONNX model by aggregating duplicate constant values as much as possible. Simple Constant value Shrink for ONNX.
Key concept
- If the same constant tensor is found by scanning the entire graph for Constant values, it is aggregated into a single constant tensor.
- Ignore scalar values.
- Ignore variables.
-
Finally, create a Fork of onnx-simplifier and merge this process just before the onnx file output process-> Temporarily abandoned because it turned out that the onnx-simplifier specification needed to be changed in a major way. - Implementation of a specification for separating the weight of a specified OP name to an external file.
- Implementation of a specification for separating the weight of a specified Constant name to an external file.
- Added option to downcast from Float64 to Float32 and INT64 to INT32 to attempt size compression.
- Final work-around idea for breaking the 2GB limit, since the internal logic of onnx has a Protocol Buffers limit of 2GB checked. Recombine after optimization. Splitting and merging seems like it would be easy. For each partitioned onnx component, optimization is performed in the order of onnx-simplifier → scs4onnx to optimize the structure while keeping the buffer size to a minimum, and then the optimized components are recombined to reconstruct the whole graph. Finally, run scs4onnx again on the reconstructed, optimized overall graph to further reduce the model-wide constant.
1. Setup
1-1. HostPC
### option
$ echo export PATH="~/.local/bin:$PATH" >> ~/.bashrc \
&& source ~/.bashrc
### run
$ pip install -U onnx \
&& python3 -m pip install -U onnx_graphsurgeon --index-url https://pypi.ngc.nvidia.com \
&& pip install -U scs4onnx
1-2. Docker
### docker pull
$ docker pull pinto0309/scs4onnx:latest
### docker build
$ docker build -t pinto0309/scs4onnx:latest .
### docker run
$ docker run --rm -it -v `pwd`:/workdir pinto0309/scs4onnx:latest
$ cd /workdir
2. CLI Usage
$ scs4onnx -h
usage:
scs4onnx [-h]
[--mode {shrink,npy}]
[--forced_extraction_op_names FORCED_EXTRACTION_OP_NAMES]
[--non_verbose]
input_onnx_file_path output_onnx_file_path
positional arguments:
input_onnx_file_path
Input onnx file path.
output_onnx_file_path
Output onnx file path.
optional arguments:
-h, --help
show this help message and exit
--mode {shrink,npy}
Constant Value Compression Mode.
shrink: Share constant values inside the model as much as possible.
The model size is slightly larger because
some shared constant values remain inside the model,
but performance is maximized.
npy: Outputs constant values used repeatedly in the model to an
external file .npy. Instead of the smallest model body size,
the file loading overhead is greater.
Default: shrink
--forced_extraction_op_names FORCED_EXTRACTION_OP_NAMES
Extracts the constant value of the specified OP name to .npy
regardless of the mode specified.
Specify the name of the OP, separated by commas.
e.g. --forced_extraction_op_names aaa,bbb,ccc
--non_verbose
Do not show all information logs. Only error logs are displayed.
3. In-script Usage
$ python
>>> from scs4onnx import shrinking
>>> help(shrinking)
Help on function shrinking in module scs4onnx.onnx_shrink_constant:
shrinking(
input_onnx_file_path: Union[str, NoneType] = '',
output_onnx_file_path: Union[str, NoneType] = '',
onnx_graph: Union[onnx.onnx_ml_pb2.ModelProto, NoneType] = None,
mode: Union[str, NoneType] = 'shrink',
forced_extraction_op_names: List[str] = [],
non_verbose: Union[bool, NoneType] = False
) -> Tuple[onnx.onnx_ml_pb2.ModelProto, str]
Parameters
----------
input_onnx_file_path: Optional[str]
Input onnx file path.
Either input_onnx_file_path or onnx_graph must be specified.
output_onnx_file_path: Optional[str]
Outpu onnx file path.
If output_onnx_file_path is not specified, no .onnx file is output.
onnx_graph: Optional[onnx.ModelProto]
onnx.ModelProto.
Either input_onnx_file_path or onnx_graph must be specified.
onnx_graph If specified, ignore input_onnx_file_path and process onnx_graph.
mode: Optional[str]
Constant Value Compression Mode.
'shrink': Share constant values inside the model as much as possible.
The model size is slightly larger because some shared constant values remain
inside the model, but performance is maximized.
'npy': Outputs constant values used repeatedly in the model to an external file .npy.
Instead of the smallest model body size, the file loading overhead is greater.
Default: shrink
forced_extraction_op_names: List[str]
Extracts the constant value of the specified OP name to .npy
regardless of the mode specified. e.g. ['aaa','bbb','ccc']
non_verbose: Optional[bool]
Do not show all information logs. Only error logs are displayed.
Default: False
Returns
-------
shrunken_graph: onnx.ModelProto
Shrunken onnx ModelProto
npy_file_paths: List[str]
List of paths to externally output .npy files.
An empty list is always returned when in 'shrink' mode.
3. CLI Execution
$ scs4onnx input.onnx output.onnx --mode shrink
4. In-script Execution
4-1. When an onnx file is used as input
If output_onnx_file_path is not specified, no .onnx file is output.
from scs4onnx import shrinking
shrunk_graph, npy_file_paths = shrinking(
input_onnx_file_path='input.onnx',
output_onnx_file_path='output.onnx',
mode='npy',
non_verbose=False
)
4-2. When entering the onnx.ModelProto
onnx_graph If specified, ignore input_onnx_file_path and process onnx_graph.
from scs4onnx import shrinking
shrunk_graph, npy_file_paths = shrinking(
onnx_graph=graph,
mode='npy',
non_verbose=True
)
5. Sample
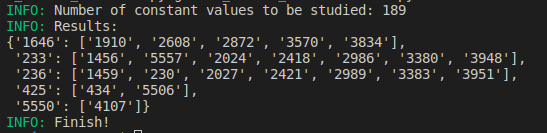
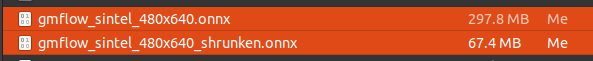
5-1. shrink mode sample
-
297.8MB -> 67.4MB
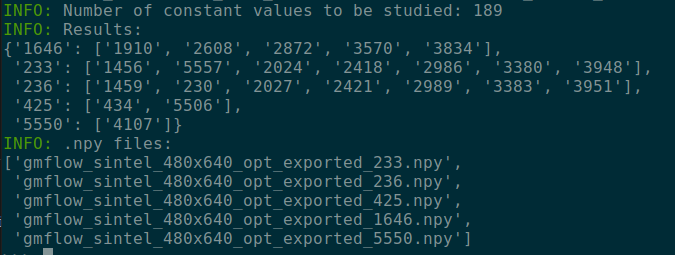
5-2. npy mode sample
-
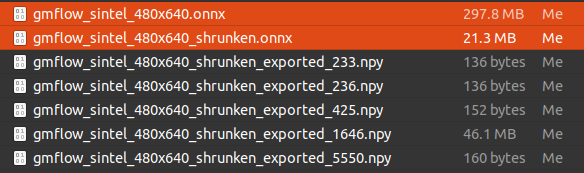
297.8MB -> 21.3MB
5-3. .npy file view
$ python
>>> import numpy as np
>>> param = np.load('gmflow_sintel_480x640_shrunken_exported_1646.npy')
>>> param.shape
(8, 1200, 1200)
>>> param
array([[[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.],
[ 0., 0., 0., ..., 0., 0., 0.],
...,
[-100., -100., -100., ..., 0., 0., 0.],
[-100., -100., -100., ..., 0., 0., 0.],
[-100., -100., -100., ..., 0., 0., 0.]]], dtype=float32)
6. Reference
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
scs4onnx-1.0.9.tar.gz
(10.2 kB
view hashes)