A RNAseq pipeline from raw reads to feature counts
Project description



This is is the RNA-seq pipeline from the Sequana project
- Overview:
RNASeq analysis from raw data to feature counts
- Input:
A set of Fastq Files and genome reference and annotation.
- Output:
MultiQC and HTML reports, BAM and bigwig files, feature Counts, script to launch differential analysis
- Status:
Production.
- Citation(sequana):
Cokelaer et al, (2017), ‘Sequana’: a Set of Snakemake NGS pipelines, Journal of Open Source Software, 2(16), 352, JOSS DOI doi:10.21105/joss.00352
- Citation(pipeline):
-
Installation
sequana_rnaseq is based on Python3, just install the package as follows:
pip install sequana_rnaseq –upgrade
You will need third-party software such as bowtie2/star. However, if you choose to use aptainer/singularity, then nothing to install except singularity itself ! See below for details.
Usage
sequana_rnaseq --help sequana_rnaseq --input-directory DATAPATH --genome-directory genome --aligner-choice star
This creates a directory with the pipeline and configuration file. You will then need to execute the pipeline:
cd rnaseq sh rnaseq.sh # for a local run
This launch a snakemake pipeline. If you are familiar with snakemake, you can retrieve the pipeline itself and its configuration files and then execute the pipeline yourself with specific parameters:
snakemake -s rnaseq.rules -c config.yaml --cores 4 --stats stats.txt
Or use sequanix interface.
Usage with apptainer:
With apptainer, initiate the working directory as follows:
sequana_rnaseq --use-apptainer
Images are downloaded in the working directory but you can store then in a directory globally (e.g.):
sequana_rnaseq --use-apptainer --apptainer-prefix ~/.sequana/apptainers
and then:
cd rnaseq sh rnaseq.sh
if you decide to use snakemake manually, do not forget to add apptainer options:
snakemake -s rnaseq.rules -c config.yaml --cores 4 --use-apptainer --apptainer-prefix ~/.sequana/apptainers --apptainer-args "-B /home:/home"
Requirements
This pipelines requires lots of third-party executable(s). Here is a list that may change. A Message will inform you would you be missing an executable:
bowtie
bowtie2>=2.4.2
STAR
featureCounts (subread package)
picard
multiqc
samtools
Note that bowtie>=2.4.2 is set to ensure the pipeline can be used with python 3.7-3.8-3.9 and the sequana-wrappers that supports bowtie2 with option –threads only (not previous versions). See environment.yaml or conda.yaml for latest list of required third-party tools.
You can install most of the tools using damona:
damona create --name sequana_tools damona activate sequana_tools damona install sequana_tools
Or use the conda.yaml file available in this repository. If you start a new environment from scratch, those commands will create the environment and install all dependencies for you:
conda create --name sequana_env python 3.7.3 conda activate sequana_env conda install -c anaconda qt pyqt>5 pip install sequana pip install sequana_rnaseq conda install --file https://raw.githubusercontent.com/sequana/rnaseq/main/conda.yaml
For Linux users, we provide singularity images available through within the damona project (https://damona.readthedocs.io).
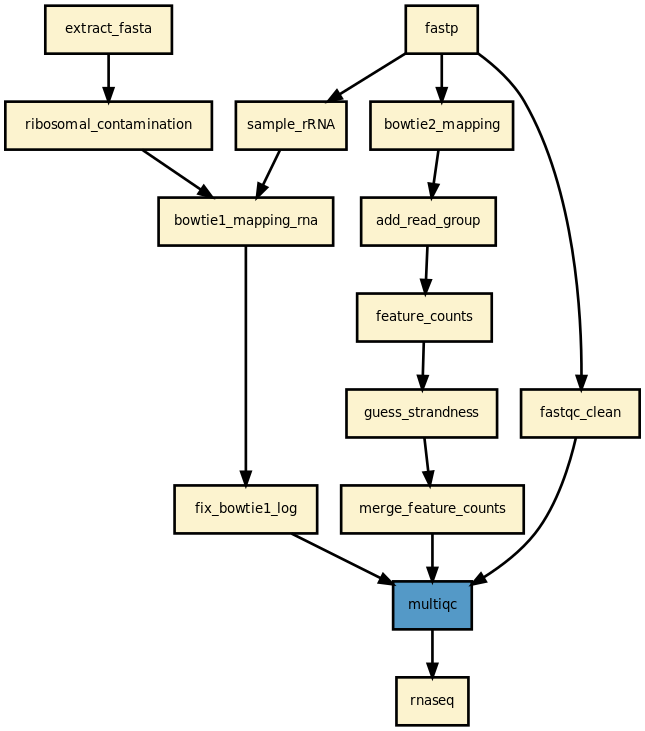
Details
This pipeline runs a RNA-seq analysis of sequencing data. It runs in parallel on a set of input FastQ files (paired or not). A brief HTML report is produced together with a MultiQC report.
This pipeline is complex and requires some expertise for the interpretation. Many online-resources are available and should help you deciphering the output.
Yet, it should be quite straigtforward to execute it as shown above. The pipeline uses bowtie1 to look for ribosomal contamination (rRNA). Then, it cleans the data with cutapdat if you say so (your data may already be pre-processed). If no adapters are provided (default), reads are trimmed for low quality bases only. Then, mapping is performed with standard mappers such as star or bowtie2 (–aligner option). Finally, feature counts are extracted from the previously generated BAM files. We guess the strand and save the feature counts into the directoy ./rnadiff/feature_counts.
The pipelines stops there. However, RNA-seq analysis are followed by a different analysis (DGE hereafter). Although the DGE is not part of the pipeline, you can performed it with standard tools using the data in ./rnadiff directory. One such tool is provided within our framework (based on the well known DEseq2 software).
Using our framework:
cd rnadiff
sequana rnadiff --design design.csv --features all_features.out --annotation ANNOT \
--feature-name FEAT --attribute-name ATTR
where ANNOT is the annotation file of your analysis, FEAT and ATTR the attribute and feature used in your analysis (coming from the annotation file).
This produces a HTML repot summarizing you differential analysis.
Rules and configuration details
Here is the latest documented configuration file to be used with the pipeline. Each rule used in the pipeline may have a section in the configuration file.
Issues
In the context of eukaryotes, you will need 32G of memory most probably. If this is too much, you can try to restrict the memory. Check out the config.yaml file in the star section.
Changelog
Version |
Description |
|---|---|
0.20.0 |
|
0.19.3 |
|
0.19.2 |
|
0.19.1 |
|
0.19.0 |
|
0.18.1 |
|
0.18.0 |
|
0.17.2 |
|
0.17.1 |
|
0.17.0 |
|
0.16.1 |
|
0.16.0 |
|
0.15.2 |
|
0.15.1 |
|
0.15.0 |
|
0.14.2 |
|
0.14.1 |
|
0.14.0 |
|
0.13.0 |
|
0.12.1 |
|
0.12.0 |
|
0.11.0 |
|
0.10.0 |
|
0.9.20 |
|
0.9.19 |
|
0.9.18 |
|
0.9.17 |
|
0.9.16 |
|
0.9.15 |
|
0.9.14 |
|
0.9.13 |
|
0.9.12 |
|
0.9.11 |
|
0.9.10 |
|
0.9.9 |
|
0.9.8 |
|
0.9.7 |
|
0.9.6 |
|
0.9.5 |
|
0.9.4 |
|
0.9.3 |
if a fastq_screen.conf is provided, we switch the fastqc_screen section ON automatically |
0.9.0 |
Major refactorisation.
|
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file sequana_rnaseq-0.20.0.tar.gz.
File metadata
- Download URL: sequana_rnaseq-0.20.0.tar.gz
- Upload date:
- Size: 81.4 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.8.2 CPython/3.10.14 Linux/6.8.9-300.fc40.x86_64
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | a689f868705a0c61e34c0a99f432a9dacf060166e8bc37d938d80a9e7cf92126 |
|
| MD5 | 25f98ceb0431a58373b2c3616d3ef89a |
|
| BLAKE2b-256 | 1a5859ffe3f081bf37b01993949b347b5c45b2a58804a22d3099fe8b89c0f959 |
Provenance
File details
Details for the file sequana_rnaseq-0.20.0-py3-none-any.whl.
File metadata
- Download URL: sequana_rnaseq-0.20.0-py3-none-any.whl
- Upload date:
- Size: 80.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: poetry/1.8.2 CPython/3.10.14 Linux/6.8.9-300.fc40.x86_64
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 433bf548073b069881ffc2db23d85e0c0a2619adaa635c01447c26460e9560d6 |
|
| MD5 | e417fb388a32306c838611fbd9fb5f3e |
|
| BLAKE2b-256 | 4b098ab7e654376a648294f69a31e8ac1a6982140659b6a2485a5869b364b1fa |













