A RNAseq pipeline from raw reads to feature counts
Project description
This is is the rnaseq pipeline from the Sequana projet
- Overview:
RNASeq analysis from raw data to feature counts
- Input:
A set of Fastq Files and genome reference and annotation.
- Output:
MultiQC reports and feature Counts
- Status:
Production
- Citation:
Cokelaer et al, (2017), ‘Sequana’: a Set of Snakemake NGS pipelines, Journal of Open Source Software, 2(16), 352, JOSS DOI doi:10.21105/joss.00352
Installation
You must install Sequana first:
pip install sequana
Then, just install this package:
pip install sequana_rnaseq
Usage
sequana_pipelines_rnaseq --help sequana_pipelines_rnaseq --input-directory DATAPATH --genome-directory --aligner star
This creates a directory with the pipeline and configuration file. You will then need to execute the pipeline:
cd rnaseq sh rnaseq.sh # for a local run
This launch a snakemake pipeline. If you are familiar with snakemake, you can retrieve the pipeline itself and its configuration files and then execute the pipeline yourself with specific parameters:
snakemake -s rnaseq.rules -c config.yaml --cores 4 --stats stats.txt
Or use sequanix interface.
Requirements
This pipelines requires the following executable(s):
bowtie
bowtie2
STAR
featureCounts
picard
multiqc
More may be needed depending on the configuration file options. For instance, you may use fastq_screen, in which case you need to install it and configure it.
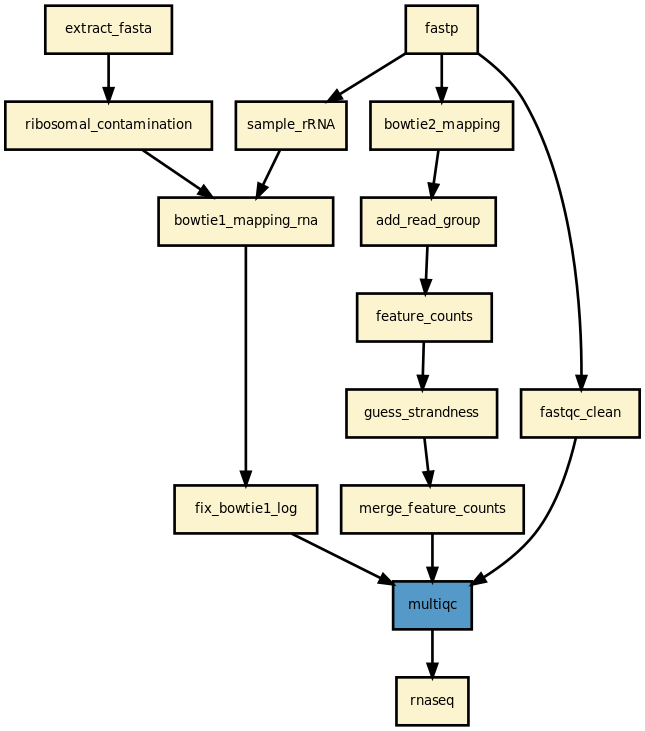
Details
This pipeline runs rnaseq in parallel on the input fastq files (paired or not). A brief sequana summary report is also produced.
Rules and configuration details
Here is the latest documented configuration file to be used with the pipeline. Each rule used in the pipeline may have a section in the configuration file.
Changelog
Version |
Description |
|---|---|
0.9.5 |
|
0.9.4 |
|
0.9.3 |
if a fastq_screen.conf is provided, we switch the fastqc_screen section ON automatically |
0.9.0 |
Major refactorisation.
|
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
File details
Details for the file sequana_rnaseq-0.9.5.tar.gz.
File metadata
- Download URL: sequana_rnaseq-0.9.5.tar.gz
- Upload date:
- Size: 87.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.13.0 pkginfo/1.4.2 requests/2.20.1 setuptools/40.6.2 requests-toolbelt/0.8.0 tqdm/4.32.2 CPython/3.6.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | bc21b449d7e5ac6e87d751531223dbb9b1d795b51b90cc218d857d2b590d7f75 |
|
| MD5 | b8020367e9e71586d1e4867f9fd2dc65 |
|
| BLAKE2b-256 | b4b15a685179e25e6be92934bd9cae3388a8333c5b9648887a92c0597ab6245f |












