Scikit-lean models hyperparameters tuning, using evolutionary algorithms
Project description

Sklearn-genetic-opt
scikit-learn models hyperparameters tuning and feature selection, using evolutionary algorithms.
This is meant to be an alternative from popular methods inside scikit-learn such as Grid Search and Randomized Grid Search for hyperparameteres tuning, and from RFE, Select From Model for feature selection.
Sklearn-genetic-opt uses evolutionary algorithms from the DEAP package to choose the set of hyperparameters that optimizes (max or min) the cross-validation scores, it can be used for both regression and classification problems.
Documentation is available here
Main Features:
GASearchCV: Main class of the package for hyperparameters tuning, holds the evolutionary cross-validation optimization routine.
GAFeatureSelectionCV: Main class of the package for feature selection.
Algorithms: Set of different evolutionary algorithms to use as an optimization procedure.
Callbacks: Custom evaluation strategies to generate early stopping rules, logging (into TensorBoard, .pkl files, etc) or your custom logic.
Plots: Generate pre-defined plots to understand the optimization process.
MLflow: Build-in integration with mlflow to log all the hyperparameters, cv-scores and the fitted models.
Demos on Features:
Visualize the progress of your training:
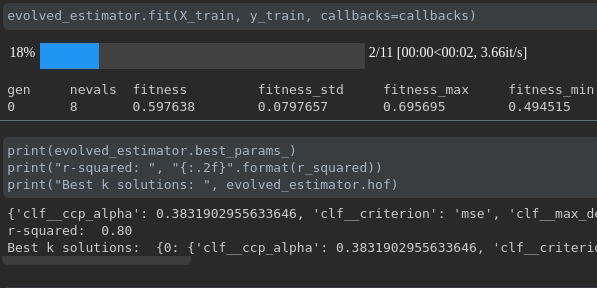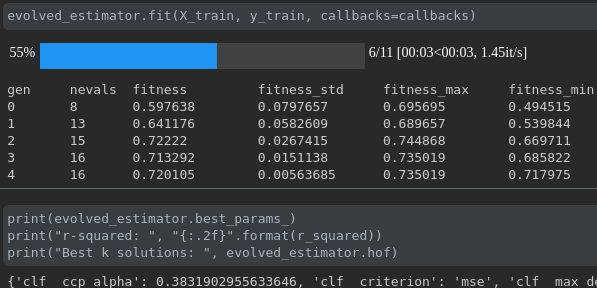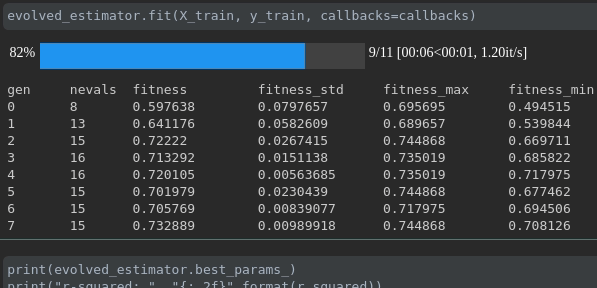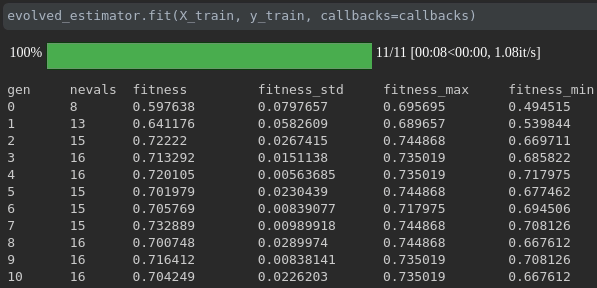
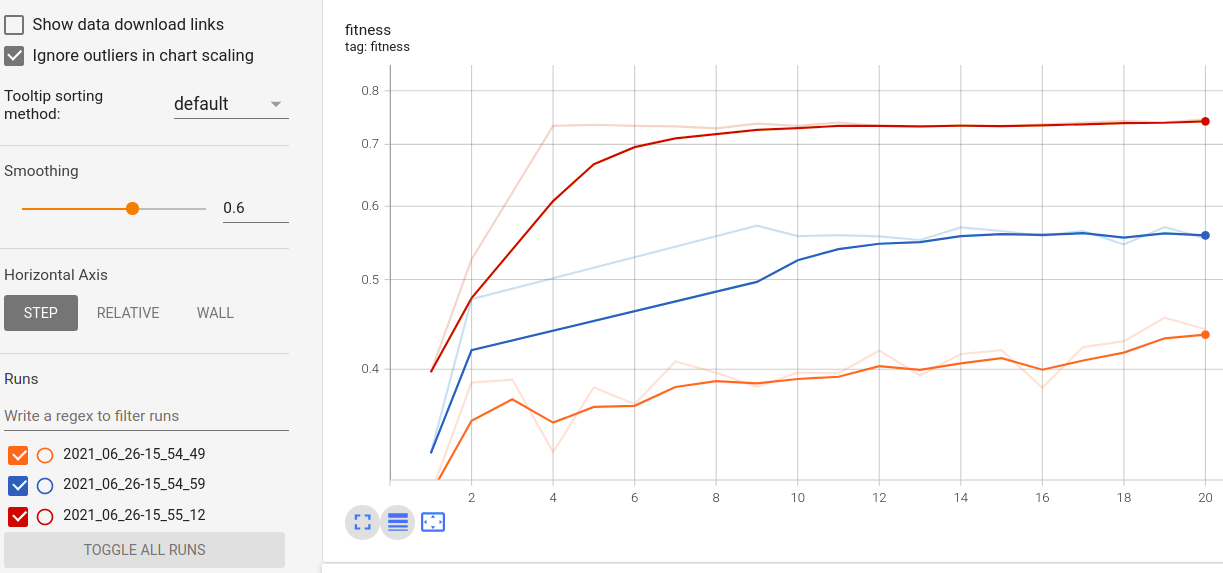
Real-time metrics visualization and comparison across runs:
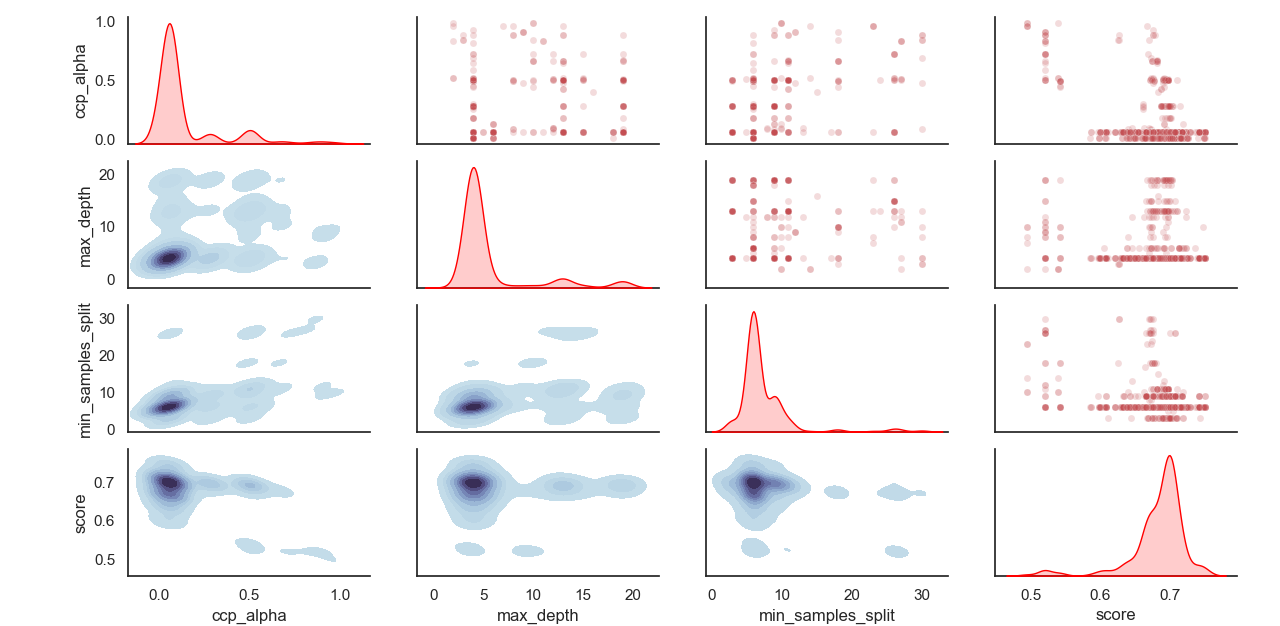
Sampled distribution of hyperparameters:
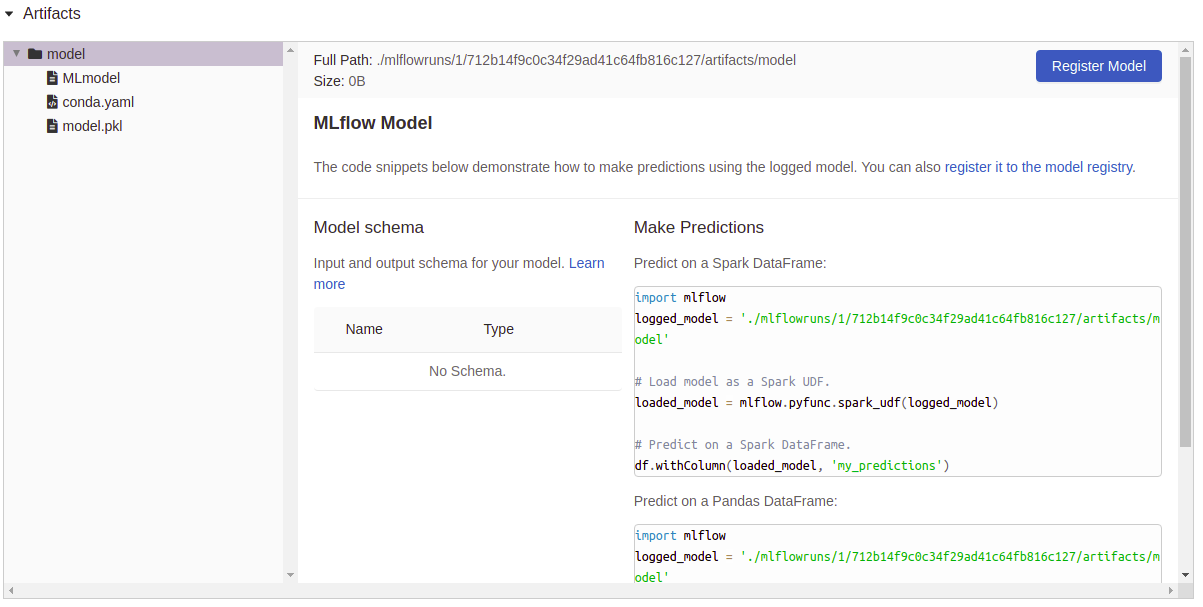
Artifacts logging:
Usage:
Install sklearn-genetic-opt
It’s advised to install sklearn-genetic using a virtual env, inside the env use:
pip install sklearn-genetic-opt
If you want to get all the features, including plotting and mlflow logging capabilities, install all the extra packages:
pip install sklearn-genetic-opt[all]
The only optional dependency that the last command does not install, it’s Tensorflow, it is usually advised to look further which distribution works better for you.
Example: Hyperparameters Tuning
from sklearn_genetic import GASearchCV
from sklearn_genetic.space import Continuous, Categorical, Integer
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split, StratifiedKFold
from sklearn.datasets import load_digits
from sklearn.metrics import accuracy_score
import matplotlib.pyplot as plt
data = load_digits()
n_samples = len(data.images)
X = data.images.reshape((n_samples, -1))
y = data['target']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)
clf = RandomForestClassifier()
param_grid = {'min_weight_fraction_leaf': Continuous(0.01, 0.5, distribution='log-uniform'),
'bootstrap': Categorical([True, False]),
'max_depth': Integer(2, 30),
'max_leaf_nodes': Integer(2, 35),
'n_estimators': Integer(100, 300)}
cv = StratifiedKFold(n_splits=3, shuffle=True)
evolved_estimator = GASearchCV(estimator=clf,
cv=cv,
scoring='accuracy',
population_size=10,
generations=35,
param_grid=param_grid,
n_jobs=-1,
verbose=True,
keep_top_k=4)
# Train and optimize the estimator
evolved_estimator.fit(X_train, y_train)
# Best parameters found
print(evolved_estimator.best_params_)
# Use the model fitted with the best parameters
y_predict_ga = evolved_estimator.predict(X_test)
print(accuracy_score(y_test, y_predict_ga))
# Saved metadata for further analysis
print("Stats achieved in each generation: ", evolved_estimator.history)
print("Best k solutions: ", evolved_estimator.hof)Example: Feature Selection
import matplotlib.pyplot as plt
from sklearn_genetic import GAFeatureSelectionCV
from sklearn.model_selection import train_test_split, StratifiedKFold
from sklearn.svm import SVC
from sklearn.datasets import load_iris
from sklearn.metrics import accuracy_score
import numpy as np
data = load_iris()
X, y = data["data"], data["target"]
# Add random non-important features
noise = np.random.uniform(0, 10, size=(X.shape[0], 5))
X = np.hstack((X, noise))
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=0)
clf = SVC(gamma='auto')
evolved_estimator = GAFeatureSelectionCV(
estimator=clf,
scoring="accuracy",
population_size=30,
generations=20,
n_jobs=-1)
# Train and select the features
evolved_estimator.fit(X_train, y_train)
# Features selected by the algorithm
features = evolved_estimator.best_features_
print(features)
# Predict only with the subset of selected features
y_predict_ga = evolved_estimator.predict(X_test[:, features])
print(accuracy_score(y_test, y_predict_ga))Changelog
See the changelog for notes on the changes of Sklearn-genetic-opt
Important links
Official source code repo: https://github.com/rodrigo-arenas/Sklearn-genetic-opt/
Download releases: https://pypi.org/project/sklearn-genetic-opt/
Issue tracker: https://github.com/rodrigo-arenas/Sklearn-genetic-opt/issues
Stable documentation: https://sklearn-genetic-opt.readthedocs.io/en/stable/
Source code
You can check the latest development version with the command:
git clone https://github.com/rodrigo-arenas/Sklearn-genetic-opt.git
Install the development dependencies:
pip install -r dev-requirements.txt
Check the latest in-development documentation: https://sklearn-genetic-opt.readthedocs.io/en/latest/
Contributing
Contributions are more than welcome! There are several opportunities on the ongoing project, so please get in touch if you would like to help out. Make sure to check the current issues and also the Contribution guide.
Big thanks to the people who are helping with this project!
Testing
After installation, you can launch the test suite from outside the source directory:
pytest sklearn_genetic
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file sklearn-genetic-opt-0.7.0.tar.gz.
File metadata
- Download URL: sklearn-genetic-opt-0.7.0.tar.gz
- Upload date:
- Size: 26.6 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.3.0 pkginfo/1.7.1 requests/2.26.0 setuptools/58.0.4 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.9.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | f9c5bbec0321893aed2ce0afc1c9817185146ae5a9c27cd14b6bcf15f6d65278 |
|
| MD5 | 79ccdd297ae606689904da962ac42869 |
|
| BLAKE2b-256 | c298d6c8c6b82318f14df788a1967eb7851e262349bca23d6df10c20561eff76 |
File details
Details for the file sklearn_genetic_opt-0.7.0-py3-none-any.whl.
File metadata
- Download URL: sklearn_genetic_opt-0.7.0-py3-none-any.whl
- Upload date:
- Size: 29.3 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.3.0 pkginfo/1.7.1 requests/2.26.0 setuptools/58.0.4 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.9.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 152571ddd54eae92d3428077a18633faee73b10ead29c6903c0c0abc61c2b3b1 |
|
| MD5 | a41e045a211d50ca5cf66a686ea0ff56 |
|
| BLAKE2b-256 | 1b6df2deb882a67323bb3b71726d5f1faa98698b18a0ca47d4a406a60e3084bb |

















