Spatial-omics pipeline and analysis
Project description

Spatial omics pipeline and analysis
Built on top of SpatialData, Sopa enables processing and analyses of spatial omics data with single-cell resolution (spatial transcriptomics or multiplex imaging data) using a standard data structure and output. We currently support the following technologies: Xenium, Visium HD, MERSCOPE, CosMx, PhenoCycler, MACSima, Molecular Cartography, and others. Sopa was designed for generability and low memory consumption on large images (scales to 1TB+ images).
[!NOTE] You may also be interested in Novae, developed by the same authors, now published in Nature Methods 🎉
Documentation
Check Sopa's documentation to get started. It contains installation explanations, CLI/API details, and tutorials.
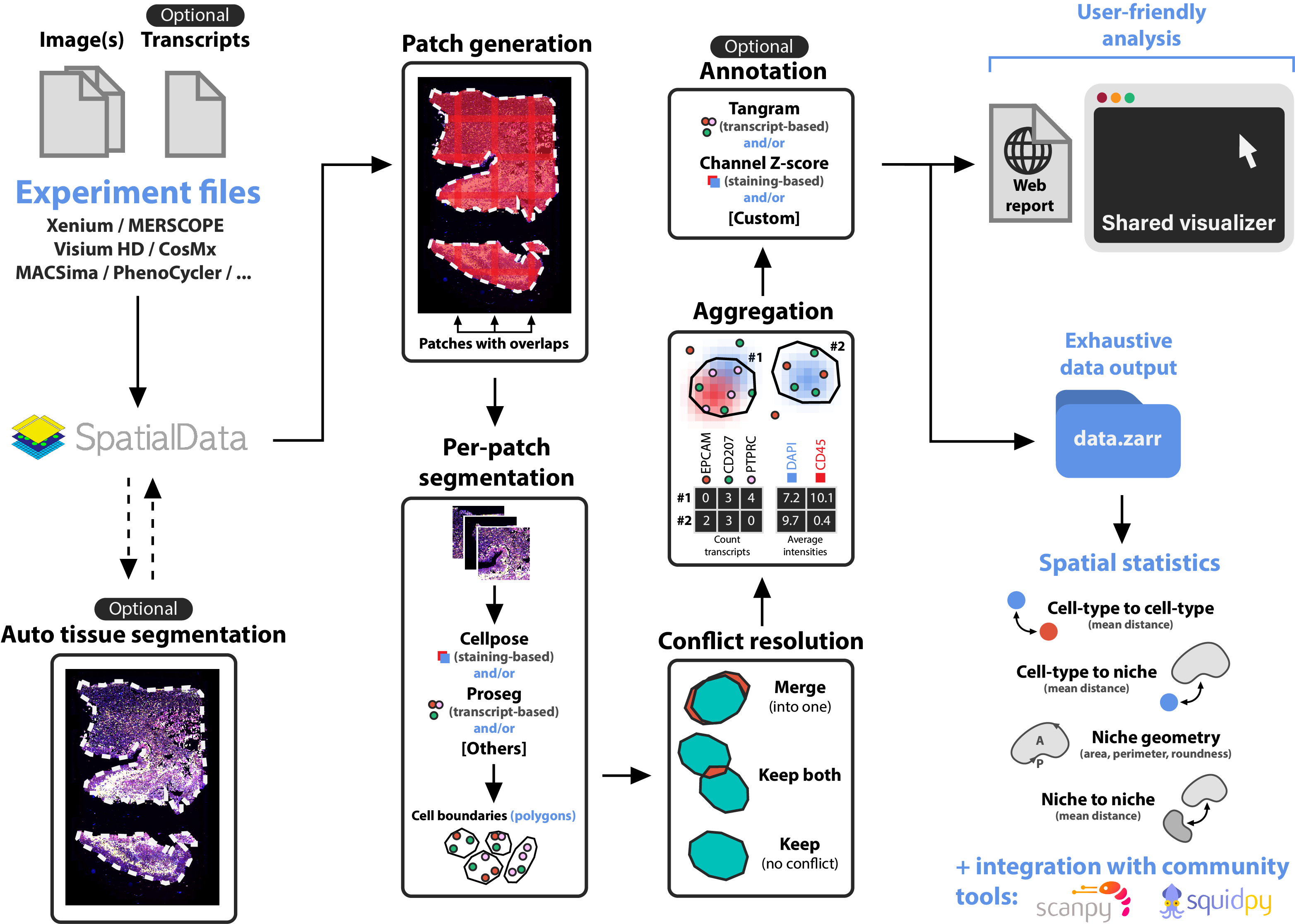
Overview
The following illustration describes the main steps of sopa:
Installation
Sopa can be installed from PyPI on all OS, for any Python version from 3.10 to 3.13 (included). On a new environment, run the following command:
pip install sopa
[!NOTE] See this installation section for more details about extras and other installations modes.
Features
Sopa comes in four different flavours, each corresponding to a different use case:
API: use directlysopaas a Python package for complete flexibility and customization.Snakemake pipeline: choose a config, and run our pipeline on your spatial data in a couple of minutes.nf-core/sopa: run Sopa with Nextflow (see this repo and the corresponding usage guide). Great for Docker users.CLI: use our command-line-interface for prototyping quickly your own pipeline (advanced users).
API
Below is a minimal example of API usage. For a complete API description, please refer to the documentation.
import sopa
sdata = sopa.io.xenium("path/to/data") # reading Xenium data
sopa.make_image_patches(sdata) # creating overlapping patches
sopa.segmentation.cellpose(sdata, "DAPI", diameter=30) # running cellpose segmentation
sopa.aggregate(sdata) # counting the transcripts inside the cells
Snakemake pipeline
Clone our repository, choose a config here (or create your own), and execute our pipeline locally or on a high-performance cluster:
git clone https://github.com/gustaveroussy/sopa.git
cd sopa/workflow
snakemake --configfile=/path/to/yaml_config --config data_path=/path/to/data_directory --cores 1 --use-conda
For more details on snakemake configuration and how to properly setup your environments, please refer to the documentation.
CLI
Below are examples of commands that can be run with the sopa CLI. For a complete description of the CLI, please refer to the documentation.
> sopa --help # show command names and arguments
> sopa convert merscope_directory --technology merscope # read some data
> sopa patchify image merscope_directory.zarr # make patches for low-memory segmentation
> sopa segmentation cellpose merscope_directory.zarr --diameter 60 --channels DAPI # segmentation
> sopa resolve cellpose merscope_directory.zarr # resolve segmentation conflicts at boundaries
> sopa aggregate merscope_directory.zarr --average-intensities # transcripts/channels aggregation
> sopa explorer write merscope_directory.zarr # convert for interactive vizualisation
Cite us
Our article is published in Nature Communications. You can cite Sopa as below:
Blampey, Q., Mulder, K., Gardet, M. et al. Sopa: a technology-invariant pipeline for analyses of image-based spatial omics.
Nat Commun 15, 4981 (2024). https://doi.org/10.1038/s41467-024-48981-z
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file sopa-2.1.11.tar.gz.
File metadata
- Download URL: sopa-2.1.11.tar.gz
- Upload date:
- Size: 138.8 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: uv/0.5.20
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
65c03377981b978a25277a9745f4a868e5ded1d2a41605f00358396203ebb83e
|
|
| MD5 |
05e5f4e0905485f771fa7b25913d1a9f
|
|
| BLAKE2b-256 |
3a0f6577bc2fe6e8ec7a1ee0342bc25d0896d546d54dc4c3dbf1fe58efc6e251
|
File details
Details for the file sopa-2.1.11-py3-none-any.whl.
File metadata
- Download URL: sopa-2.1.11-py3-none-any.whl
- Upload date:
- Size: 182.2 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: uv/0.5.20
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
0d3b89bec97729b895e1bf1d55211513f8c682ba1c7ae27b7b0d8363034243eb
|
|
| MD5 |
1ea914c811b4972ae5e09c4bb7c3a3ae
|
|
| BLAKE2b-256 |
881eeac4f166cfa4d0ad4058164ccbd2e87096c2eb766b97103851dbcbaee5fa
|


















