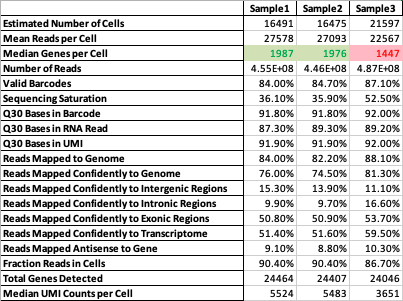
Easily assess quality control data across multiple single cell datasets
Project description
Easily assess quality control data across multiple single cell datasets
Supercells is a tool which generates a single report for multiple 10x cellranger samples - thus saving time and allowing comparative QC analysis.
It is written in Python (tested with v3.7-3.9).
📦 Installation
Using PyPI:
pip install supercells
Clone or download the repository and navigate to the package directory, then invoke:
pip install .
💻 Usage
Once supercells is installed it is run by invoking:
supercells -i <experiment folder>
Unless specified otherwise it will generate supercells_report.html within the input folder as well as supercells_report folder containing summarizing xlsx file and CSV with the raw data. To specify an output directory use the -o <output folder> flag
More options
usage: supercells [-h] [--version] --input INPATH [--output OUTPATH] [--cutoff-dict CUTOFF_DICT_PATH]
supercells: Easily assess quality control data across multiple single cell datasets
options:
-h, --help show this help message and exit
--version, -v show program's version number and exit
--input INPATH, -i INPATH
Enter the location of the input files
--output OUTPATH, -o OUTPATH
Specify the location of the output files, default is current wd
--cutoff-dict CUTOFF_DICT_PATH, -c CUTOFF_DICT_PATH
cutoff-dict parameters is a file path to a json file.
This is a sample of the json:
{
"Mean Reads per Cell": 20000,
"Median Genes per Cell": 1500,
"Valid Barcodes": "75%",
"Sequencing Saturation": "30%",
"Q30 Bases in RNA Read": 65,
"Fraction Reads in Cells": "70%",
"Reads Mapped to Genome": "75%",
"Reads Mapped Confidently to Transcriptome": "30%"
}
The keys are the field names and the values are cutoff values for that field. i.e. Mean Reads per Cell cutoff
value is 20000. So for each sample if the Mean Reads per Cell is less than 20000 the output would show
that cell in red, else in green.
Further development
Suggestions for additional features and code contributions are welcomed
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distributions
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file supercells-0.0.5-py3-none-any.whl.
File metadata
- Download URL: supercells-0.0.5-py3-none-any.whl
- Upload date:
- Size: 9.9 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.1 CPython/3.9.12
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
7b881907f316edb426f82d1451a63d4deff296969bfca1749e6d19e9da82d947
|
|
| MD5 |
fcdfb82bbda6e0f59ce51e41db3d2e8c
|
|
| BLAKE2b-256 |
792fe2f161985bde8743129ba4a0860a0b196b0dd0466c0339bf9ff95c24f10a
|