No project description provided
Project description
synthaser
Process
synthaser parses the results of a batch NCBI conserved domain search and determines
the domain architecture of secondary metabolite synthases.
Installation
Install from PyPI via pip:
$ pip install synthaser
or clone the repo and install locally:
$ git clone https://www.github.com/gamcil/synthaser
$ cd synthaser
$ pip install -e .
Dependencies
synthaser is written for Python 3.6+ and has been tested on Linux (Ubuntu 18.04) and
Windows (10). The only external Python dependency is requests, which is used for
querying the NCBI's APIs.
Usage
A search can be launched as simply as:
$ synthaser -qi <accessions> OR synthaser -qf query.fasta
For example, performing a synthaser run on the cichorine PKS:
$ synthaser -qi CBF69451.1
[11:13:42] INFO - Starting synthaser
[11:13:44] INFO - Launching new CDSearch run on IDs: ['CBF69451.1']
[11:13:45] INFO - Run ID: QM3-qcdsearch-14C5BC063AA03DDE-15B11AB00918AED0
[11:13:45] INFO - Polling NCBI for results...
[11:13:45] INFO - Checking search status...
[11:14:05] INFO - Checking search status...
[11:14:06] INFO - Search successfully completed!
NR-PKS
------
CBF69451.1 SAT-KS-AT-PT-ACP-ACP-MT-TE
[11:14:06] INFO - Finished synthaser
synthaser can also produce an SVG representation of the query synthases. For example,
we could take the CDSID (CD-Search ID) of the previous run, and provide the --svg flag:
$ synthaser -qi CBF69451.1 \
--cdsid QM3-qcdsearch-14C5BC063AA03DDE-15B11AB00918AED0 \
--svg figure.svg
The generated figure is then saved in figure.svg, and looks like:
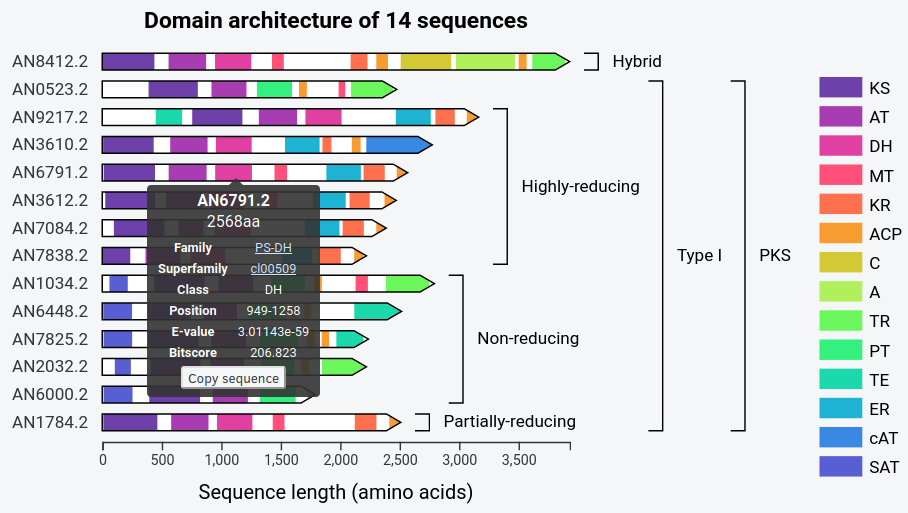
synthaser can also start batch searches, either by providing more than one sequence in
a query FASTA file (-qf), or more than one NCBI accession (-qi).
For example, searching PKS sequences from A. nidulans:
$ synthaser -qf sequences.fasta --json nidulans.svg
Produces:
Refer to synthaser --help for all tweakable parameters for generating the SVG.
Citations
If you found synthaser helpful, please cite:
1. <pending>
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
Filter files by name, interpreter, ABI, and platform.
If you're not sure about the file name format, learn more about wheel file names.
Copy a direct link to the current filters
File details
Details for the file synthaser-1.0.2.tar.gz.
File metadata
- Download URL: synthaser-1.0.2.tar.gz
- Upload date:
- Size: 29.1 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.14.0 pkginfo/1.5.0.1 requests/2.22.0 setuptools/39.0.1 requests-toolbelt/0.9.1 tqdm/4.35.0 CPython/3.6.8
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
c1f1d017824ea7748d796748f2b05a6255e4ccda3662f9b89c4b50e45f66a009
|
|
| MD5 |
2047a576efc6c6d2d8952efc6cf21062
|
|
| BLAKE2b-256 |
1ce02cd3d4c8659e551d1196657530bc8c041bf73e692f0076ab0e604f859adb
|
File details
Details for the file synthaser-1.0.2-py3-none-any.whl.
File metadata
- Download URL: synthaser-1.0.2-py3-none-any.whl
- Upload date:
- Size: 36.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.14.0 pkginfo/1.5.0.1 requests/2.22.0 setuptools/39.0.1 requests-toolbelt/0.9.1 tqdm/4.35.0 CPython/3.6.8
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 |
8bb4c058e1d4321c7d05a3e6574498c0d632abbb82a1931593a562c575202f22
|
|
| MD5 |
64c509c0de0b170a6c6abc423fff41e5
|
|
| BLAKE2b-256 |
e7113dad706607bd9c69798c5730aa7b8c4780f32e51cf9691a18aa2f3efe9e8
|