A novel method for unsupervised patient stratification.
Project description
UnPaSt
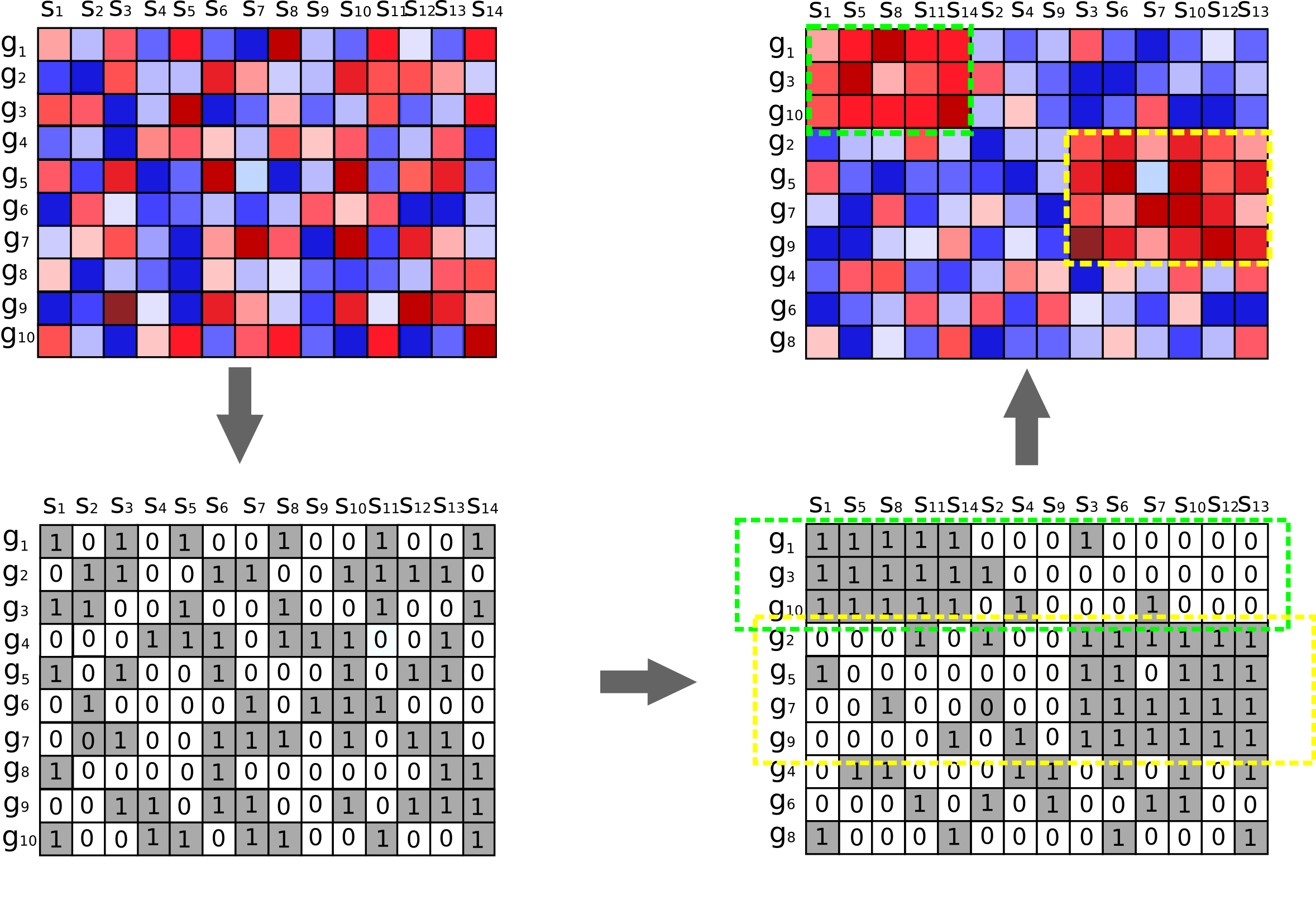
UnPaSt is a novel method for identification of differentially expressed biclusters in gene expression matrix. It searches for gene sets up- or down-regulated in subsets of samples:
Webserver: https://unpast.zbh.uni-hamburg.de/
Installation
This UnPaSt can be installed using pip./poster/DESMOND2.png, poetry, run using Docker, or as a script (see examples section). Follow the appropriate instructions below for your preferred method.
You need to have R and Python 3.8 installed.
- Using pip:
To install the project usingpip, first make sure you havepipinstalled on your system. If you haven't installed it already, you can find the installation instructions here.
Oncepipis installed, you can install the project by running the following command:
pip install unpast
Run it:
run_unpast -h
Dependencies. To use this package, you will need to have R and the WGCNA library installed. You can easily install these dependencies by running the following command after installing unpast:
python -m unpast.install_r_dependencies
- Installation using Poetry:
To install the package using Poetry, first make sure you have Poetry installed, clone the repo and then run:
poetry add unpast
Run it:
poetry run run_unpast -h
Dependencies. To use this package, you will need to have R and the WGCNA library installed. You can easily install these dependencies by running the following command after installing unpast:
poetry run python -m unpast.install_r_dependencies
- Running with Docker:
You can also run the package using Docker. First, pull the Docker image:
docker pull freddsle/unpast:latest
Next, run the container:
docker run -v /your/data/path/:/user_data/ freddsle/unpast:latest --exprs /user_data/exprs.tsv --out_dir /user_data/out_dir/
Examples
- UnPaSt requires a tab-separated file with standardized expressions of genes (or transcripts) in rows, and samples in columns. Gene and sample names must be unique.
- A subset of 200 randomly chosen samples from TCGA-BRCA and UnPaSt output: test data
# running UnPaSt with default parameters and example data python ./unpast/run_unpast.py --exprs TCGA_200.exprs_z.tsv --basename TCGA_200_results # with different binarization and clustering methods python ./unpast/run_unpast.py --exprs TCGA_200.exprs_z.tsv --basename results --binarization ward --clustering WGCNA # help python ./unpast/run_unpast.py -h
Outputs
- <basename>.bin=[GMM|Jenks],clust=[Louvain|WGCNA|DESMOND].biclusters.tsv - a .tsv table with found biclsuters, where
- avgSNR is average SNR over all genes in the biclusters
- columns "n_genes" and "n_samples" provide the numbers of genes and samples, respectively
- "gene","sample" contain gene and sample names respectively
- "gene_indexes" and "sample_indexes" - 0-based gene and sample indexes in the input matrix.
- binarized expressions, background distributions of SNR for each bicluster size and binarization statistics [if clustering is WGCNA, or '--save_binary' flag is added]
- modules found by WGCNA [if clustering is WGCNA]
About
UnPaSt is an unconstrained version of DESMOND method (repository, publication)
Major modifications:
- it does not require the network of gene interactions
- UnPaSt clusters individual genes instead of gene pairs
- uses Gaussian mixture models or Jenks method for binarization of individual gene expressions
- SNR threshold is authomatically determined; it depends on bicluster size in samples and user-defined p-value cutoff
License
Free for non-for-profit use. For commercial use please contact the developers.
Poster CDCS workshop'22
Poster ISMB and MCCMB'21
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.